Caroline Uhler
Laboratory for Information & Decision Systems, Massachusetts Institute of Technology
Latent Causal Diffusions for Single-Cell Perturbation Modeling
Jan 20, 2026Abstract:Perturbation screens hold the potential to systematically map regulatory processes at single-cell resolution, yet modeling and predicting transcriptome-wide responses to perturbations remains a major computational challenge. Existing methods often underperform simple baselines, fail to disentangle measurement noise from biological signal, and provide limited insight into the causal structure governing cellular responses. Here, we present the latent causal diffusion (LCD), a generative model that frames single-cell gene expression as a stationary diffusion process observed under measurement noise. LCD outperforms established approaches in predicting the distributional shifts of unseen perturbation combinations in single-cell RNA-sequencing screens while simultaneously learning a mechanistic dynamical system of gene regulation. To interpret these learned dynamics, we develop an approach we call causal linearization via perturbation responses (CLIPR), which yields an approximation of the direct causal effects between all genes modeled by the diffusion. CLIPR provably identifies causal effects under a linear drift assumption and recovers causal structure in both simulated systems and a genome-wide perturbation screen, where it clusters genes into coherent functional modules and resolves causal relationships that standard differential expression analysis cannot. The LCD-CLIPR framework bridges generative modeling with causal inference to predict unseen perturbation effects and map the underlying regulatory mechanisms of the transcriptome.
Causal Structure and Representation Learning with Biomedical Applications
Nov 06, 2025



Abstract:Massive data collection holds the promise of a better understanding of complex phenomena and, ultimately, better decisions. Representation learning has become a key driver of deep learning applications, as it allows learning latent spaces that capture important properties of the data without requiring any supervised annotations. Although representation learning has been hugely successful in predictive tasks, it can fail miserably in causal tasks including predicting the effect of a perturbation/intervention. This calls for a marriage between representation learning and causal inference. An exciting opportunity in this regard stems from the growing availability of multi-modal data (observational and perturbational, imaging-based and sequencing-based, at the single-cell level, tissue-level, and organism-level). We outline a statistical and computational framework for causal structure and representation learning motivated by fundamental biomedical questions: how to effectively use observational and perturbational data to perform causal discovery on observed causal variables; how to use multi-modal views of the system to learn causal variables; and how to design optimal perturbations.
BindEnergyCraft: Casting Protein Structure Predictors as Energy-Based Models for Binder Design
May 27, 2025Abstract:Protein binder design has been transformed by hallucination-based methods that optimize structure prediction confidence metrics, such as the interface predicted TM-score (ipTM), via backpropagation. However, these metrics do not reflect the statistical likelihood of a binder-target complex under the learned distribution and yield sparse gradients for optimization. In this work, we propose a method to extract such likelihoods from structure predictors by reinterpreting their confidence outputs as an energy-based model (EBM). By leveraging the Joint Energy-based Modeling (JEM) framework, we introduce pTMEnergy, a statistical energy function derived from predicted inter-residue error distributions. We incorporate pTMEnergy into BindEnergyCraft (BECraft), a design pipeline that maintains the same optimization framework as BindCraft but replaces ipTM with our energy-based objective. BECraft outperforms BindCraft, RFDiffusion, and ESM3 across multiple challenging targets, achieving higher in silico binder success rates while reducing structural clashes. Furthermore, pTMEnergy establishes a new state-of-the-art in structure-based virtual screening tasks for miniprotein and RNA aptamer binders.
Meta-Dependence in Conditional Independence Testing
Apr 17, 2025



Abstract:Constraint-based causal discovery algorithms utilize many statistical tests for conditional independence to uncover networks of causal dependencies. These approaches to causal discovery rely on an assumed correspondence between the graphical properties of a causal structure and the conditional independence properties of observed variables, known as the causal Markov condition and faithfulness. Finite data yields an empirical distribution that is "close" to the actual distribution. Across these many possible empirical distributions, the correspondence to the graphical properties can break down for different conditional independencies, and multiple violations can occur at the same time. We study this "meta-dependence" between conditional independence properties using the following geometric intuition: each conditional independence property constrains the space of possible joint distributions to a manifold. The "meta-dependence" between conditional independences is informed by the position of these manifolds relative to the true probability distribution. We provide a simple-to-compute measure of this meta-dependence using information projections and consolidate our findings empirically using both synthetic and real-world data.
No Foundations without Foundations -- Why semi-mechanistic models are essential for regulatory biology
Jan 31, 2025Abstract:Despite substantial efforts, deep learning has not yet delivered a transformative impact on elucidating regulatory biology, particularly in the realm of predicting gene expression profiles. Here, we argue that genuine "foundation models" of regulatory biology will remain out of reach unless guided by frameworks that integrate mechanistic insight with principled experimental design. We present one such ground-up, semi-mechanistic framework that unifies perturbation-based experimental designs across both in vitro and in vivo CRISPR screens, accounting for differentiating and non-differentiating cellular systems. By revealing previously unrecognised assumptions in published machine learning methods, our approach clarifies links with popular techniques such as variational autoencoders and structural causal models. In practice, this framework suggests a modified loss function that we demonstrate can improve predictive performance, and further suggests an error analysis that informs batching strategies. Ultimately, since cellular regulation emerges from innumerable interactions amongst largely uncharted molecular components, we contend that systems-level understanding cannot be achieved through structural biology alone. Instead, we argue that real progress will require a first-principles perspective on how experiments capture biological phenomena, how data are generated, and how these processes can be reflected in more faithful modelling architectures.
Learning Mixtures of Unknown Causal Interventions
Oct 31, 2024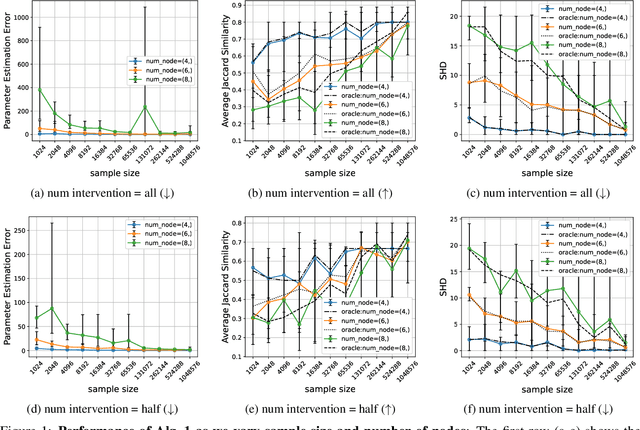
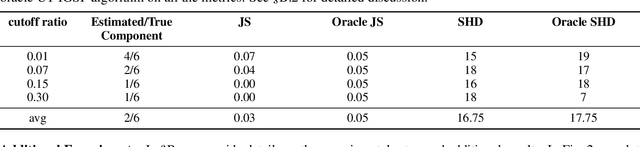
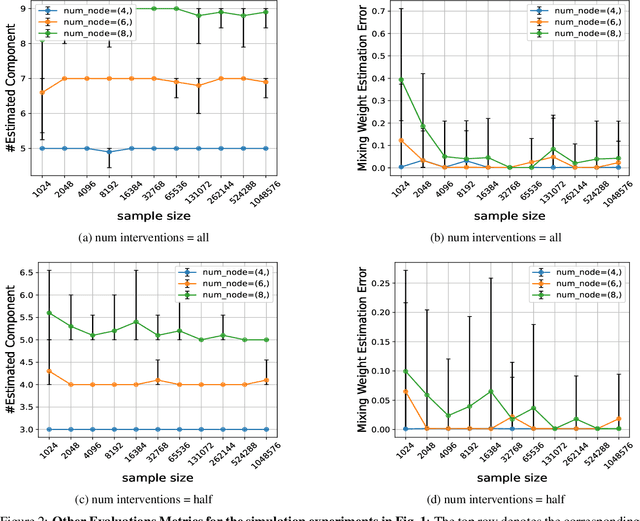
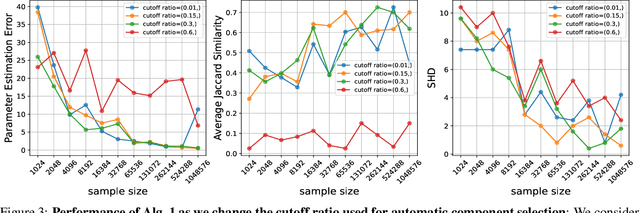
Abstract:The ability to conduct interventions plays a pivotal role in learning causal relationships among variables, thus facilitating applications across diverse scientific disciplines such as genomics, economics, and machine learning. However, in many instances within these applications, the process of generating interventional data is subject to noise: rather than data being sampled directly from the intended interventional distribution, interventions often yield data sampled from a blend of both intended and unintended interventional distributions. We consider the fundamental challenge of disentangling mixed interventional and observational data within linear Structural Equation Models (SEMs) with Gaussian additive noise without the knowledge of the true causal graph. We demonstrate that conducting interventions, whether do or soft, yields distributions with sufficient diversity and properties conducive to efficiently recovering each component within the mixture. Furthermore, we establish that the sample complexity required to disentangle mixed data inversely correlates with the extent of change induced by an intervention in the equations governing the affected variable values. As a result, the causal graph can be identified up to its interventional Markov Equivalence Class, similar to scenarios where no noise influences the generation of interventional data. We further support our theoretical findings by conducting simulations wherein we perform causal discovery from such mixed data.
An Information Criterion for Controlled Disentanglement of Multimodal Data
Oct 31, 2024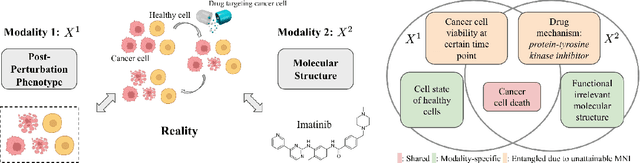
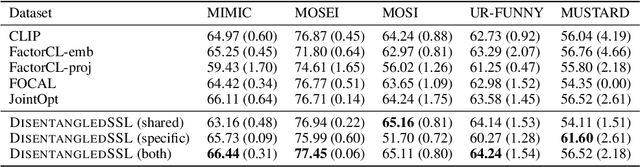
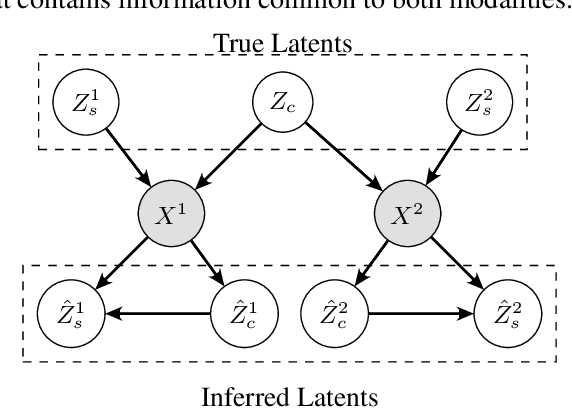
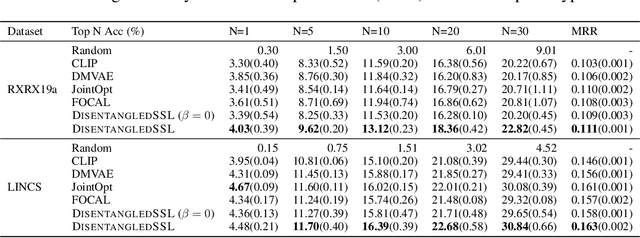
Abstract:Multimodal representation learning seeks to relate and decompose information inherent in multiple modalities. By disentangling modality-specific information from information that is shared across modalities, we can improve interpretability and robustness and enable downstream tasks such as the generation of counterfactual outcomes. Separating the two types of information is challenging since they are often deeply entangled in many real-world applications. We propose Disentangled Self-Supervised Learning (DisentangledSSL), a novel self-supervised approach for learning disentangled representations. We present a comprehensive analysis of the optimality of each disentangled representation, particularly focusing on the scenario not covered in prior work where the so-called Minimum Necessary Information (MNI) point is not attainable. We demonstrate that DisentangledSSL successfully learns shared and modality-specific features on multiple synthetic and real-world datasets and consistently outperforms baselines on various downstream tasks, including prediction tasks for vision-language data, as well as molecule-phenotype retrieval tasks for biological data.
Causal Discovery with Fewer Conditional Independence Tests
Jun 03, 2024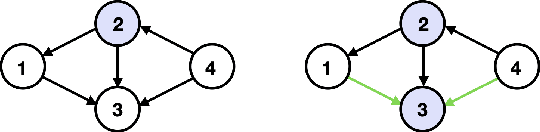


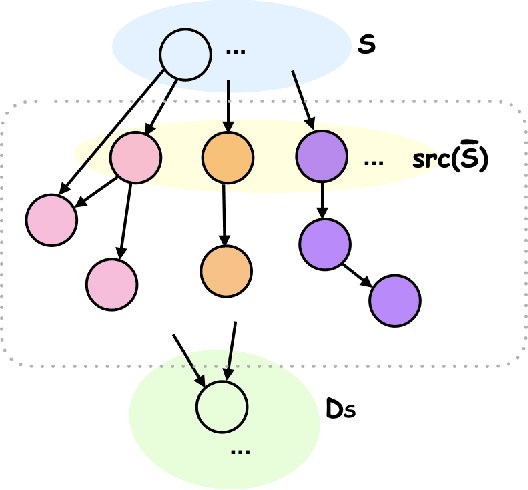
Abstract:Many questions in science center around the fundamental problem of understanding causal relationships. However, most constraint-based causal discovery algorithms, including the well-celebrated PC algorithm, often incur an exponential number of conditional independence (CI) tests, posing limitations in various applications. Addressing this, our work focuses on characterizing what can be learned about the underlying causal graph with a reduced number of CI tests. We show that it is possible to a learn a coarser representation of the hidden causal graph with a polynomial number of tests. This coarser representation, named Causal Consistent Partition Graph (CCPG), comprises of a partition of the vertices and a directed graph defined over its components. CCPG satisfies consistency of orientations and additional constraints which favor finer partitions. Furthermore, it reduces to the underlying causal graph when the causal graph is identifiable. As a consequence, our results offer the first efficient algorithm for recovering the true causal graph with a polynomial number of tests, in special cases where the causal graph is fully identifiable through observational data and potentially additional interventions.
Synthetic Potential Outcomes for Mixtures of Treatment Effects
May 29, 2024Abstract:Modern data analysis frequently relies on the use of large datasets, often constructed as amalgamations of diverse populations or data-sources. Heterogeneity across these smaller datasets constitutes two major challenges for causal inference: (1) the source of each sample can introduce latent confounding between treatment and effect, and (2) diverse populations may respond differently to the same treatment, giving rise to heterogeneous treatment effects (HTEs). The issues of latent confounding and HTEs have been studied separately but not in conjunction. In particular, previous works only report the conditional average treatment effect (CATE) among similar individuals (with respect to the measured covariates). CATEs cannot resolve mixtures of potential treatment effects driven by latent heterogeneity, which we call mixtures of treatment effects (MTEs). Inspired by method of moment approaches to mixture models, we propose "synthetic potential outcomes" (SPOs). Our new approach deconfounds heterogeneity while also guaranteeing the identifiability of MTEs. This technique bypasses full recovery of a mixture, which significantly simplifies its requirements for identifiability. We demonstrate the efficacy of SPOs on synthetic data.
Season combinatorial intervention predictions with Salt & Peper
Apr 25, 2024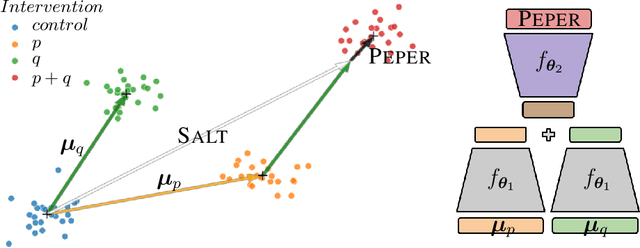
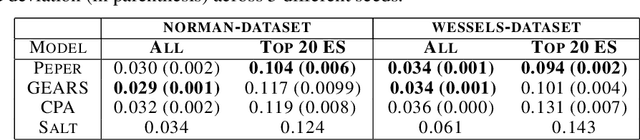
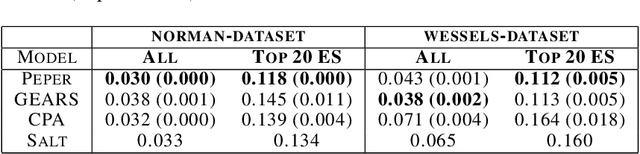
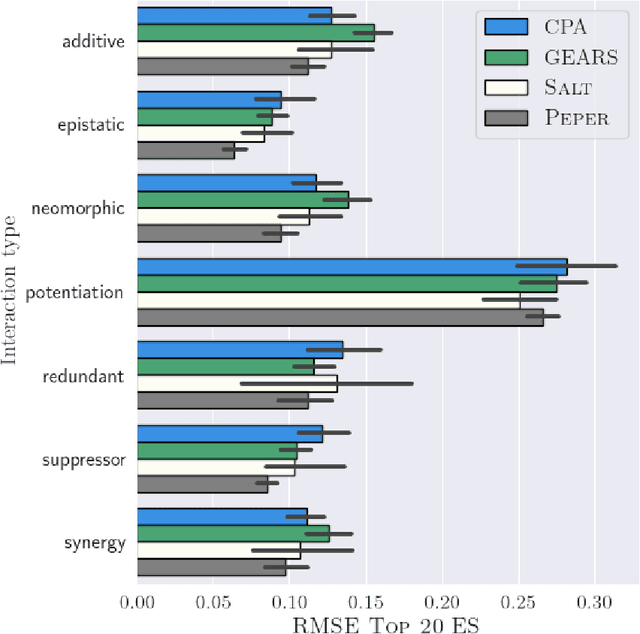
Abstract:Interventions play a pivotal role in the study of complex biological systems. In drug discovery, genetic interventions (such as CRISPR base editing) have become central to both identifying potential therapeutic targets and understanding a drug's mechanism of action. With the advancement of CRISPR and the proliferation of genome-scale analyses such as transcriptomics, a new challenge is to navigate the vast combinatorial space of concurrent genetic interventions. Addressing this, our work concentrates on estimating the effects of pairwise genetic combinations on the cellular transcriptome. We introduce two novel contributions: Salt, a biologically-inspired baseline that posits the mostly additive nature of combination effects, and Peper, a deep learning model that extends Salt's additive assumption to achieve unprecedented accuracy. Our comprehensive comparison against existing state-of-the-art methods, grounded in diverse metrics, and our out-of-distribution analysis highlight the limitations of current models in realistic settings. This analysis underscores the necessity for improved modelling techniques and data acquisition strategies, paving the way for more effective exploration of genetic intervention effects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge