Luka Kovačević
Large Language Models for Zero-shot Inference of Causal Structures in Biology
Mar 06, 2025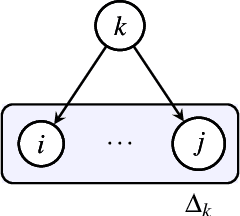
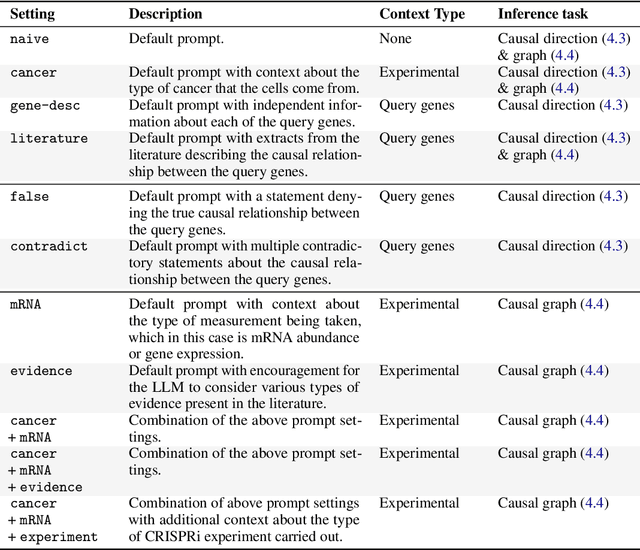
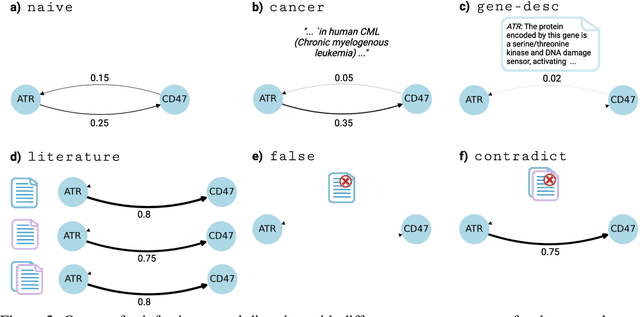
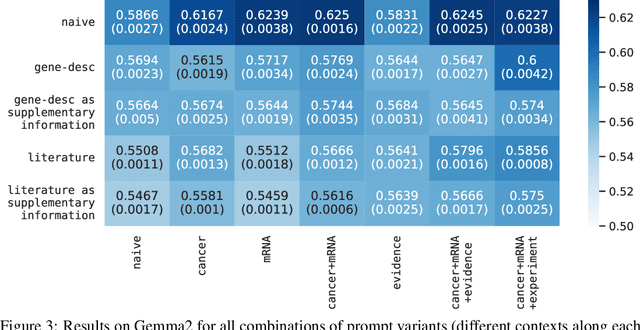
Abstract:Genes, proteins and other biological entities influence one another via causal molecular networks. Causal relationships in such networks are mediated by complex and diverse mechanisms, through latent variables, and are often specific to cellular context. It remains challenging to characterise such networks in practice. Here, we present a novel framework to evaluate large language models (LLMs) for zero-shot inference of causal relationships in biology. In particular, we systematically evaluate causal claims obtained from an LLM using real-world interventional data. This is done over one hundred variables and thousands of causal hypotheses. Furthermore, we consider several prompting and retrieval-augmentation strategies, including large, and potentially conflicting, collections of scientific articles. Our results show that with tailored augmentation and prompting, even relatively small LLMs can capture meaningful aspects of causal structure in biological systems. This supports the notion that LLMs could act as orchestration tools in biological discovery, by helping to distil current knowledge in ways amenable to downstream analysis. Our approach to assessing LLMs with respect to experimental data is relevant for a broad range of problems at the intersection of causal learning, LLMs and scientific discovery.
No Foundations without Foundations -- Why semi-mechanistic models are essential for regulatory biology
Jan 31, 2025Abstract:Despite substantial efforts, deep learning has not yet delivered a transformative impact on elucidating regulatory biology, particularly in the realm of predicting gene expression profiles. Here, we argue that genuine "foundation models" of regulatory biology will remain out of reach unless guided by frameworks that integrate mechanistic insight with principled experimental design. We present one such ground-up, semi-mechanistic framework that unifies perturbation-based experimental designs across both in vitro and in vivo CRISPR screens, accounting for differentiating and non-differentiating cellular systems. By revealing previously unrecognised assumptions in published machine learning methods, our approach clarifies links with popular techniques such as variational autoencoders and structural causal models. In practice, this framework suggests a modified loss function that we demonstrate can improve predictive performance, and further suggests an error analysis that informs batching strategies. Ultimately, since cellular regulation emerges from innumerable interactions amongst largely uncharted molecular components, we contend that systems-level understanding cannot be achieved through structural biology alone. Instead, we argue that real progress will require a first-principles perspective on how experiments capture biological phenomena, how data are generated, and how these processes can be reflected in more faithful modelling architectures.
Simulation-based Benchmarking for Causal Structure Learning in Gene Perturbation Experiments
Jul 08, 2024



Abstract:Causal structure learning (CSL) refers to the task of learning causal relationships from data. Advances in CSL now allow learning of causal graphs in diverse application domains, which has the potential to facilitate data-driven causal decision-making. Real-world CSL performance depends on a number of $\textit{context-specific}$ factors, including context-specific data distributions and non-linear dependencies, that are important in practical use-cases. However, our understanding of how to assess and select CSL methods in specific contexts remains limited. To address this gap, we present $\textit{CausalRegNet}$, a multiplicative effect structural causal model that allows for generating observational and interventional data incorporating context-specific properties, with a focus on the setting of gene perturbation experiments. Using real-world gene perturbation data, we show that CausalRegNet generates accurate distributions and scales far better than current simulation frameworks. We illustrate the use of CausalRegNet in assessing CSL methods in the context of interventional experiments in biology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge