Sach Mukherjee
Evolving Afferent Architectures: Biologically-inspired Models for Damage-Avoidance Learning
Feb 04, 2026Abstract:We introduce Afferent Learning, a framework that produces Computational Afferent Traces (CATs) as adaptive, internal risk signals for damage-avoidance learning. Inspired by biological systems, the framework uses a two-level architecture: evolutionary optimization (outer loop) discovers afferent sensing architectures that enable effective policy learning, while reinforcement learning (inner loop) trains damage-avoidance policies using these signals. This formalizes afferent sensing as providing an inductive bias for efficient learning: architectures are selected based on their ability to enable effective learning (rather than directly minimizing damage). We provide theoretical convergence guarantees under smoothness and bounded-noise assumptions. We illustrate the general approach in the challenging context of biomechanical digital twins operating over long time horizons (multiple decades of the life-course). Here, we find that CAT-based evolved architectures achieve significantly higher efficiency and better age-robustness than hand-designed baselines, enabling policies that exhibit age-dependent behavioral adaptation (23% reduction in high-risk actions). Ablation studies validate CAT signals, evolution, and predictive discrepancy as essential. We release code and data for reproducibility.
Many Experiments, Few Repetitions, Unpaired Data, and Sparse Effects: Is Causal Inference Possible?
Jan 21, 2026Abstract:We study the problem of estimating causal effects under hidden confounding in the following unpaired data setting: we observe some covariates $X$ and an outcome $Y$ under different experimental conditions (environments) but do not observe them jointly; we either observe $X$ or $Y$. Under appropriate regularity conditions, the problem can be cast as an instrumental variable (IV) regression with the environment acting as a (possibly high-dimensional) instrument. When there are many environments but only a few observations per environment, standard two-sample IV estimators fail to be consistent. We propose a GMM-type estimator based on cross-fold sample splitting of the instrument-covariate sample and prove that it is consistent as the number of environments grows but the sample size per environment remains constant. We further extend the method to sparse causal effects via $\ell_1$-regularized estimation and post-selection refitting.
Large Language Models for Zero-shot Inference of Causal Structures in Biology
Mar 06, 2025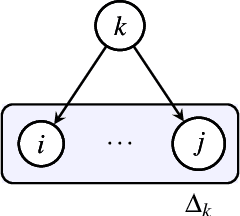
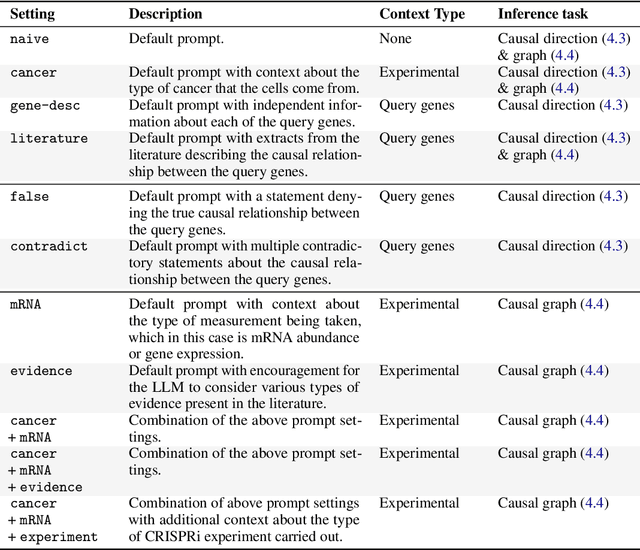
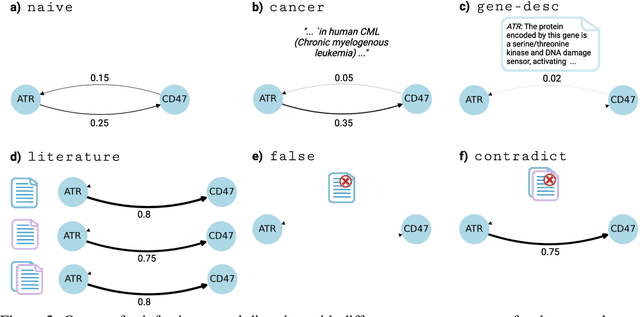
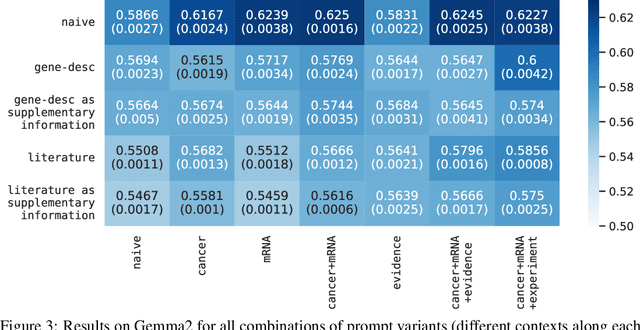
Abstract:Genes, proteins and other biological entities influence one another via causal molecular networks. Causal relationships in such networks are mediated by complex and diverse mechanisms, through latent variables, and are often specific to cellular context. It remains challenging to characterise such networks in practice. Here, we present a novel framework to evaluate large language models (LLMs) for zero-shot inference of causal relationships in biology. In particular, we systematically evaluate causal claims obtained from an LLM using real-world interventional data. This is done over one hundred variables and thousands of causal hypotheses. Furthermore, we consider several prompting and retrieval-augmentation strategies, including large, and potentially conflicting, collections of scientific articles. Our results show that with tailored augmentation and prompting, even relatively small LLMs can capture meaningful aspects of causal structure in biological systems. This supports the notion that LLMs could act as orchestration tools in biological discovery, by helping to distil current knowledge in ways amenable to downstream analysis. Our approach to assessing LLMs with respect to experimental data is relevant for a broad range of problems at the intersection of causal learning, LLMs and scientific discovery.
Simulation-based Benchmarking for Causal Structure Learning in Gene Perturbation Experiments
Jul 08, 2024



Abstract:Causal structure learning (CSL) refers to the task of learning causal relationships from data. Advances in CSL now allow learning of causal graphs in diverse application domains, which has the potential to facilitate data-driven causal decision-making. Real-world CSL performance depends on a number of $\textit{context-specific}$ factors, including context-specific data distributions and non-linear dependencies, that are important in practical use-cases. However, our understanding of how to assess and select CSL methods in specific contexts remains limited. To address this gap, we present $\textit{CausalRegNet}$, a multiplicative effect structural causal model that allows for generating observational and interventional data incorporating context-specific properties, with a focus on the setting of gene perturbation experiments. Using real-world gene perturbation data, we show that CausalRegNet generates accurate distributions and scales far better than current simulation frameworks. We illustrate the use of CausalRegNet in assessing CSL methods in the context of interventional experiments in biology.
Learning Latent Dynamics via Invariant Decomposition and (Spatio-)Temporal Transformers
Jun 21, 2023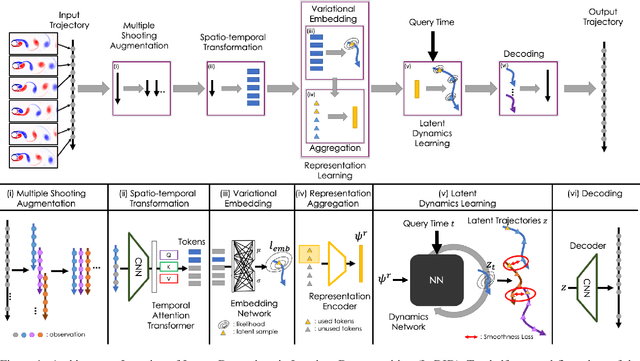
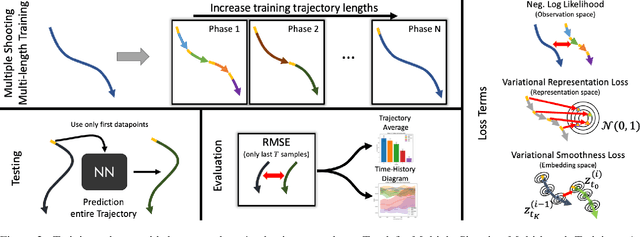
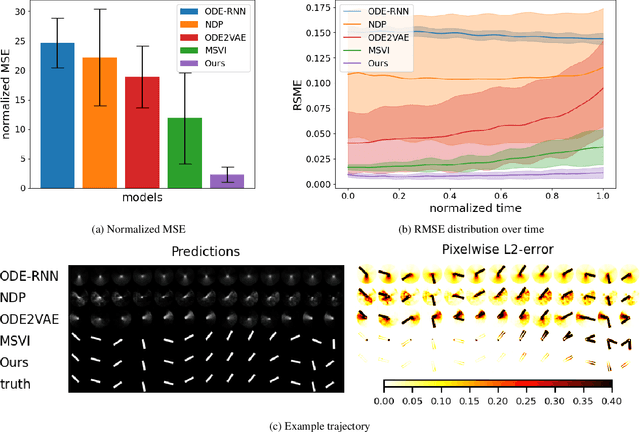
Abstract:We propose a method for learning dynamical systems from high-dimensional empirical data that combines variational autoencoders and (spatio-)temporal attention within a framework designed to enforce certain scientifically-motivated invariances. We focus on the setting in which data are available from multiple different instances of a system whose underlying dynamical model is entirely unknown at the outset. The approach rests on a separation into an instance-specific encoding (capturing initial conditions, constants etc.) and a latent dynamics model that is itself universal across all instances/realizations of the system. The separation is achieved in an automated, data-driven manner and only empirical data are required as inputs to the model. The approach allows effective inference of system behaviour at any continuous time but does not require an explicit neural ODE formulation, which makes it efficient and highly scalable. We study behaviour through simple theoretical analyses and extensive experiments on synthetic and real-world datasets. The latter investigate learning the dynamics of complex systems based on finite data and show that the proposed approach can outperform state-of-the-art neural-dynamical models. We study also more general inductive bias in the context of transfer to data obtained under entirely novel system interventions. Overall, our results provide a promising new framework for efficiently learning dynamical models from heterogeneous data with potential applications in a wide range of fields including physics, medicine, biology and engineering.
Deep Learning of Causal Structures in High Dimensions
Dec 09, 2022Abstract:Recent years have seen rapid progress at the intersection between causality and machine learning. Motivated by scientific applications involving high-dimensional data, in particular in biomedicine, we propose a deep neural architecture for learning causal relationships between variables from a combination of empirical data and prior causal knowledge. We combine convolutional and graph neural networks within a causal risk framework to provide a flexible and scalable approach. Empirical results include linear and nonlinear simulations (where the underlying causal structures are known and can be directly compared against), as well as a real biological example where the models are applied to high-dimensional molecular data and their output compared against entirely unseen validation experiments. These results demonstrate the feasibility of using deep learning approaches to learn causal networks in large-scale problems spanning thousands of variables.
High-Dimensional Undirected Graphical Models for Arbitrary Mixed Data
Nov 21, 2022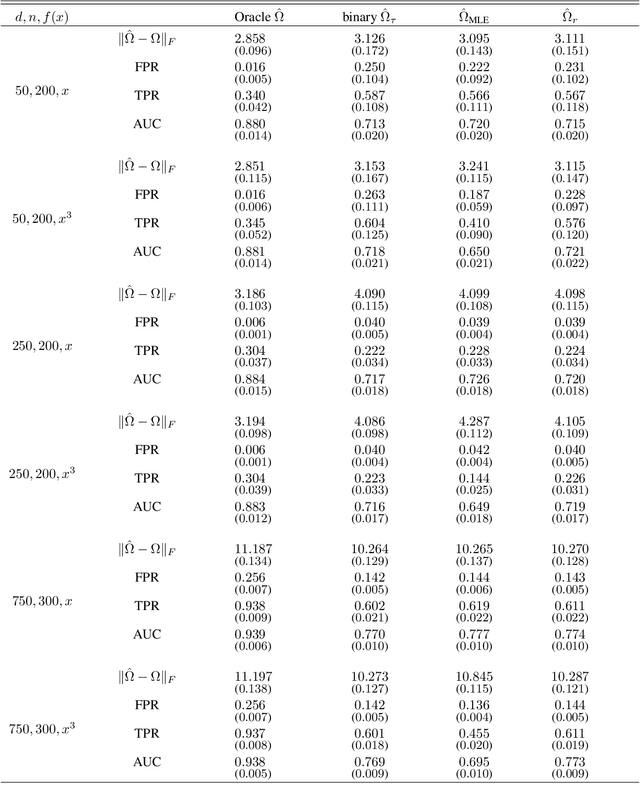
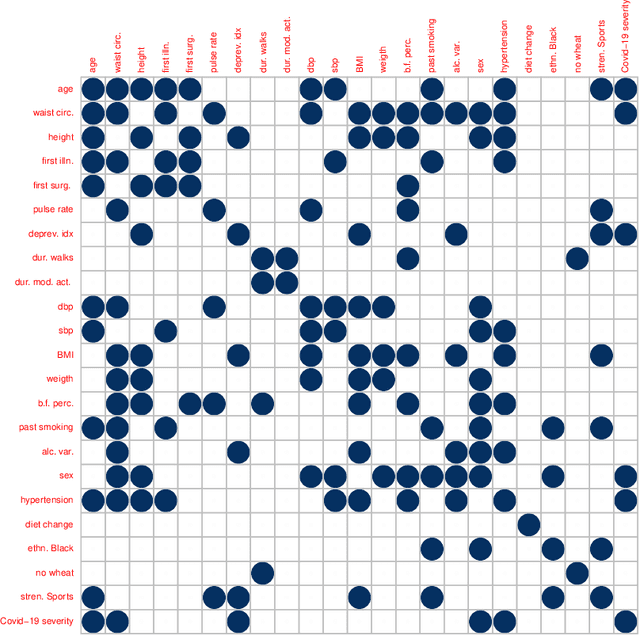
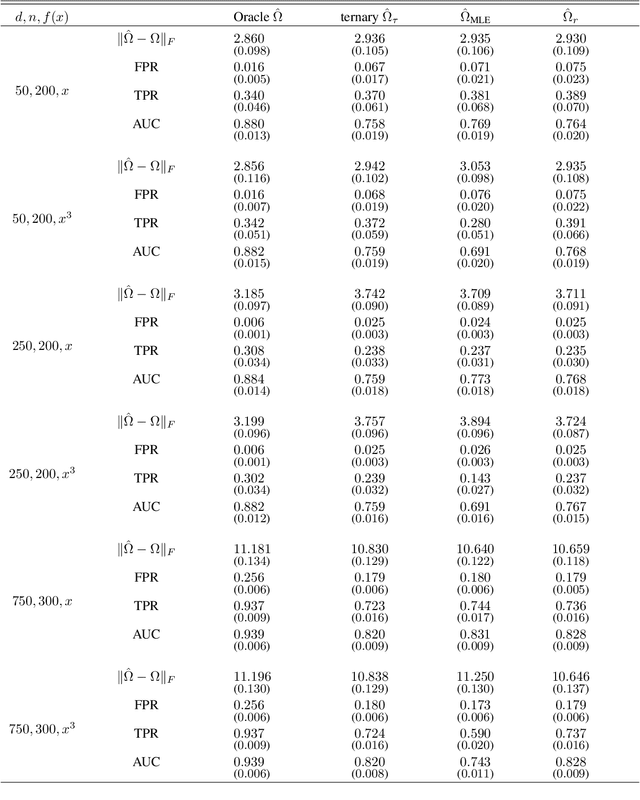
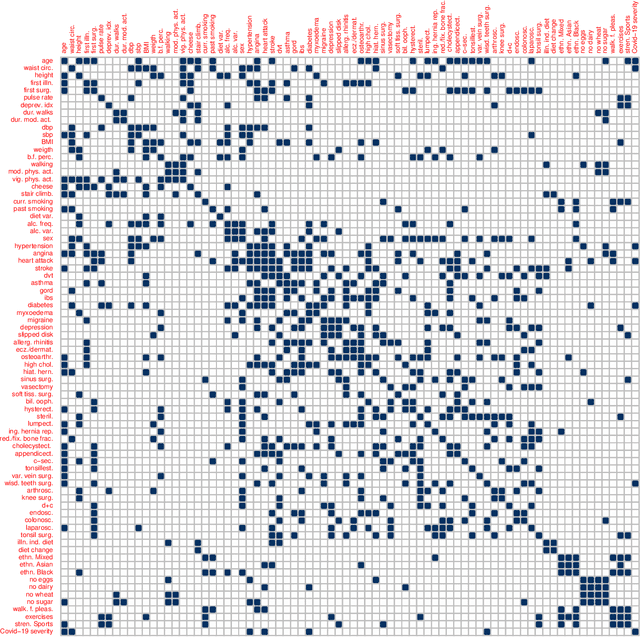
Abstract:Graphical models are an important tool in exploring relationships between variables in complex, multivariate data. Methods for learning such graphical models are well developed in the case where all variables are either continuous or discrete, including in high-dimensions. However, in many applications data span variables of different types (e.g. continuous, count, binary, ordinal, etc.), whose principled joint analysis is nontrivial. Latent Gaussian copula models, in which all variables are modeled as transformations of underlying jointly Gaussian variables, represent a useful approach. Recent advances have shown how the binary-continuous case can be tackled, but the general mixed variable type regime remains challenging. In this work, we make the simple yet useful observation that classical ideas concerning polychoric and polyserial correlations can be leveraged in a latent Gaussian copula framework. Building on this observation we propose flexible and scalable methodology for data with variables of entirely general mixed type. We study the key properties of the approaches theoretically and empirically, via extensive simulations as well an illustrative application to data from the UK Biobank concerning COVID-19 risk factors.
Scalable Regularised Joint Mixture Models
May 03, 2022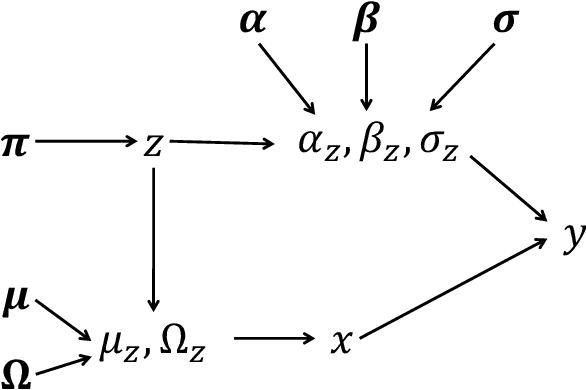
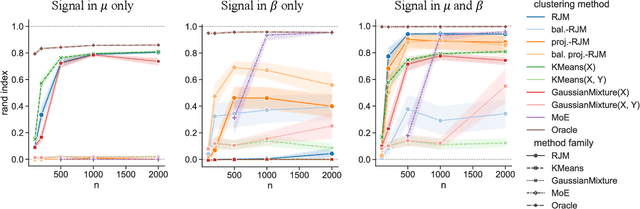
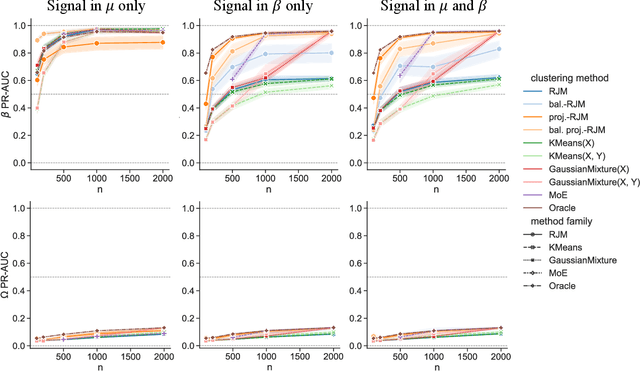
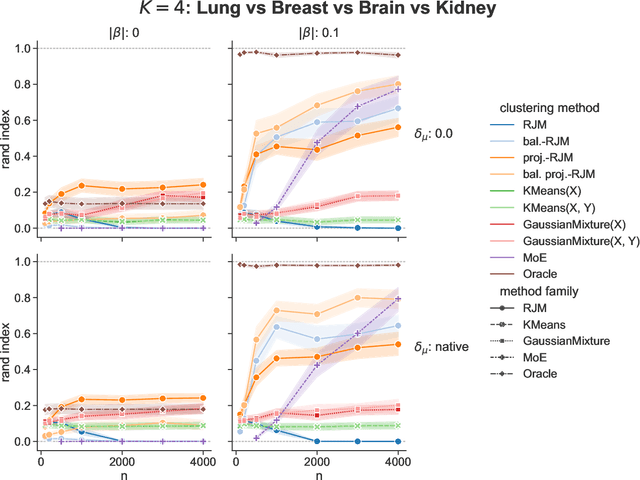
Abstract:In many applications, data can be heterogeneous in the sense of spanning latent groups with different underlying distributions. When predictive models are applied to such data the heterogeneity can affect both predictive performance and interpretability. Building on developments at the intersection of unsupervised learning and regularised regression, we propose an approach for heterogeneous data that allows joint learning of (i) explicit multivariate feature distributions, (ii) high-dimensional regression models and (iii) latent group labels, with both (i) and (ii) specific to latent groups and both elements informing (iii). The approach is demonstrably effective in high dimensions, combining data reduction for computational efficiency with a re-weighting scheme that retains key signals even when the number of features is large. We discuss in detail these aspects and their impact on modelling and computation, including EM convergence. The approach is modular and allows incorporation of data reductions and high-dimensional estimators that are suitable for specific applications. We show results from extensive simulations and real data experiments, including highly non-Gaussian data. Our results allow efficient, effective analysis of high-dimensional data in settings, such as biomedicine, where both interpretable prediction and explicit feature space models are needed but hidden heterogeneity may be a concern.
On unsupervised projections and second order signals
Apr 11, 2022
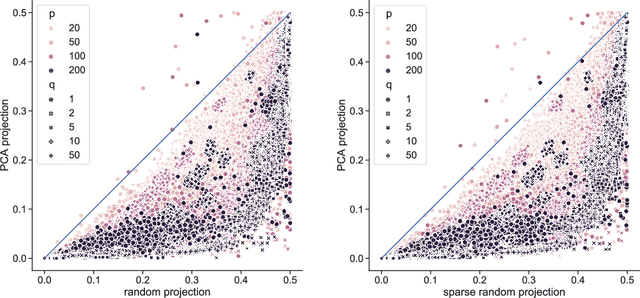
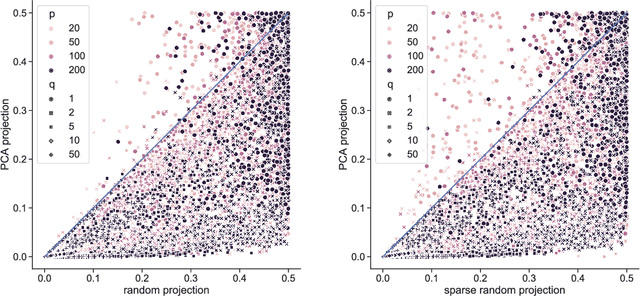
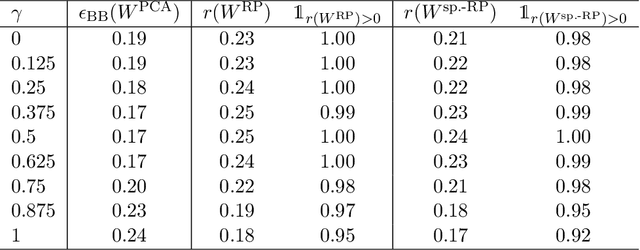
Abstract:Linear projections are widely used in the analysis of high-dimensional data. In unsupervised settings where the data harbour latent classes/clusters, the question of whether class discriminatory signals are retained under projection is crucial. In the case of mean differences between classes, this question has been well studied. However, in many contemporary applications, notably in biomedicine, group differences at the level of covariance or graphical model structure are important. Motivated by such applications, in this paper we ask whether linear projections can preserve differences in second order structure between latent groups. We focus on unsupervised projections, which can be computed without knowledge of class labels. We discuss a simple theoretical framework to study the behaviour of such projections which we use to inform an analysis via quasi-exhaustive enumeration. This allows us to consider the performance, over more than a hundred thousand sets of data-generating population parameters, of two popular projections, namely random projections (RP) and Principal Component Analysis (PCA). Across this broad range of regimes, PCA turns out to be more effective at retaining second order signals than RP and is often even competitive with supervised projection. We complement these results with fully empirical experiments showing 0-1 loss using simulated and real data. We study also the effect of projection dimension, drawing attention to a bias-variance trade-off in this respect. Our results show that PCA can indeed be a suitable first-step for unsupervised analysis, including in cases where differential covariance or graphical model structure are of interest.
Ancestral causal learning in high dimensions with a human genome-wide application
May 27, 2019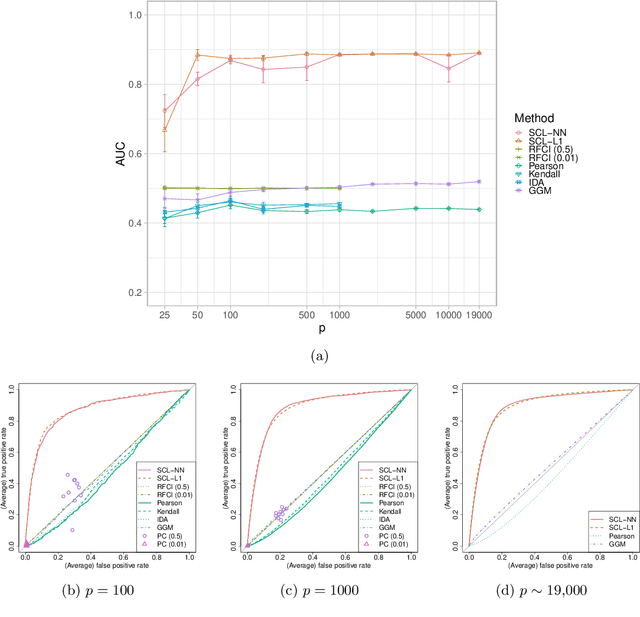
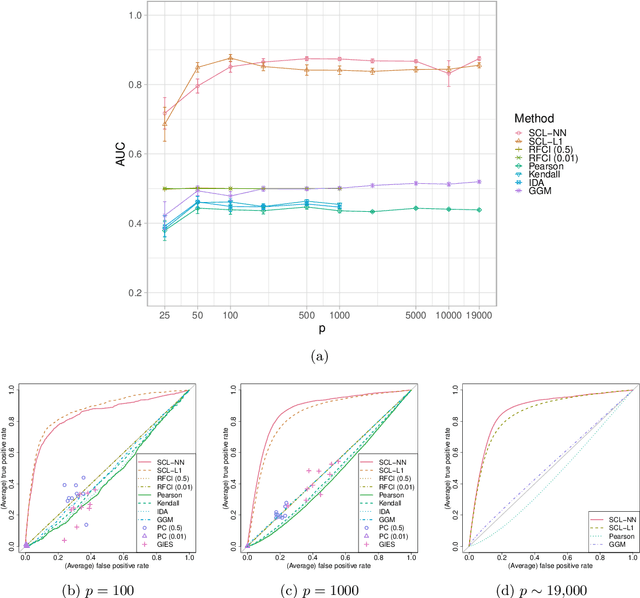
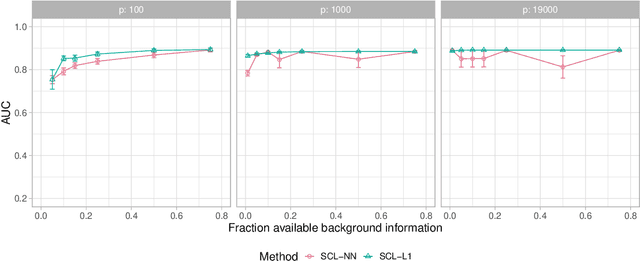
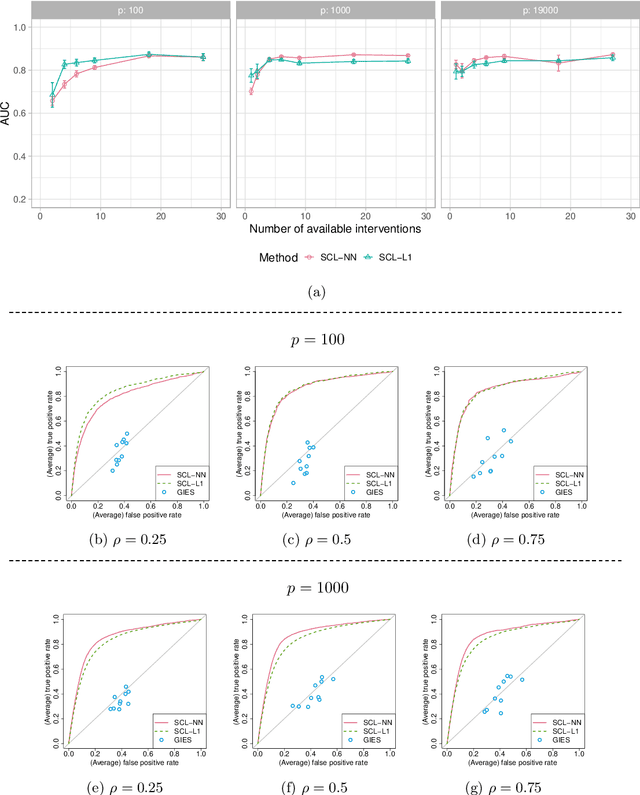
Abstract:We consider learning ancestral causal relationships in high dimensions. Our approach is driven by a supervised learning perspective, with discrete indicators of causal relationships treated as labels to be learned from available data. We focus on the setting in which some causal (ancestral) relationships are known (via background knowledge or experimental data) and put forward a general approach that scales to large problems. This is motivated by problems in human biology which are characterized by high dimensionality and potentially many latent variables. We present a case study involving interventional data from human cells with total dimension $p \! \sim \! 19{,}000$. Performance is assessed empirically by testing model output against previously unseen interventional data. The proposed approach is highly effective and demonstrably scalable to the human genome-wide setting. We consider sensitivity to background knowledge and find that results are robust to nontrivial perturbations of the input information. We consider also the case, relevant to some applications, where the only prior information available concerns a small number of known ancestral relationships.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge