Scalable Regularised Joint Mixture Models
Paper and Code
May 03, 2022
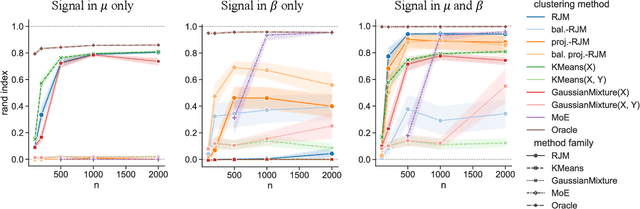
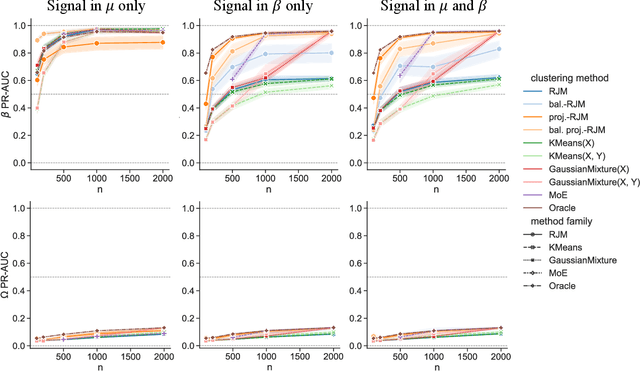
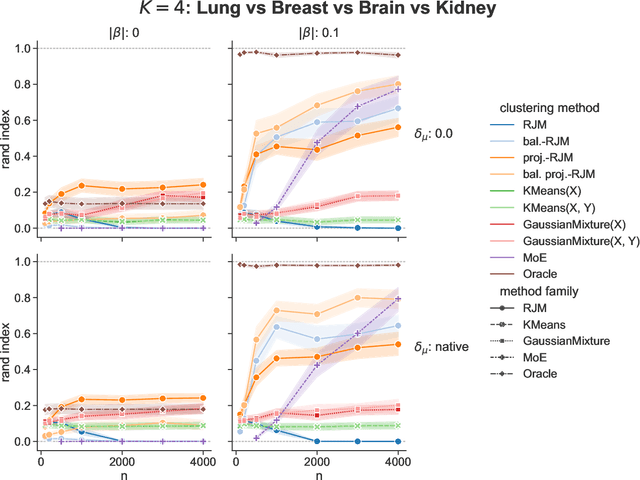
In many applications, data can be heterogeneous in the sense of spanning latent groups with different underlying distributions. When predictive models are applied to such data the heterogeneity can affect both predictive performance and interpretability. Building on developments at the intersection of unsupervised learning and regularised regression, we propose an approach for heterogeneous data that allows joint learning of (i) explicit multivariate feature distributions, (ii) high-dimensional regression models and (iii) latent group labels, with both (i) and (ii) specific to latent groups and both elements informing (iii). The approach is demonstrably effective in high dimensions, combining data reduction for computational efficiency with a re-weighting scheme that retains key signals even when the number of features is large. We discuss in detail these aspects and their impact on modelling and computation, including EM convergence. The approach is modular and allows incorporation of data reductions and high-dimensional estimators that are suitable for specific applications. We show results from extensive simulations and real data experiments, including highly non-Gaussian data. Our results allow efficient, effective analysis of high-dimensional data in settings, such as biomedicine, where both interpretable prediction and explicit feature space models are needed but hidden heterogeneity may be a concern.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge