Sylvia Richardson
Federated Variational Inference for Bayesian Mixture Models
Feb 18, 2025



Abstract:We present a federated learning approach for Bayesian model-based clustering of large-scale binary and categorical datasets. We introduce a principled 'divide and conquer' inference procedure using variational inference with local merge and delete moves within batches of the data in parallel, followed by 'global' merge moves across batches to find global clustering structures. We show that these merge moves require only summaries of the data in each batch, enabling federated learning across local nodes without requiring the full dataset to be shared. Empirical results on simulated and benchmark datasets demonstrate that our method performs well in comparison to existing clustering algorithms. We validate the practical utility of the method by applying it to large scale electronic health record (EHR) data.
Bayesian outcome-guided multi-view mixture models with applications in molecular precision medicine
Mar 01, 2023

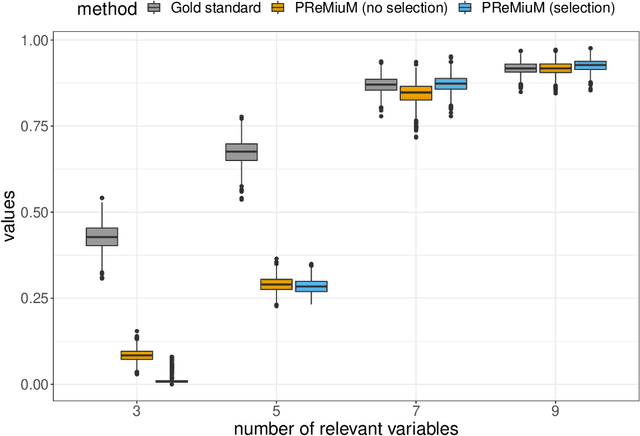

Abstract:Clustering is commonly performed as an initial analysis step for uncovering structure in 'omics datasets, e.g. to discover molecular subtypes of disease. The high-throughput, high-dimensional nature of these datasets means that they provide information on a diverse array of different biomolecular processes and pathways. Different groups of variables (e.g. genes or proteins) will be implicated in different biomolecular processes, and hence undertaking analyses that are limited to identifying just a single clustering partition of the whole dataset is therefore liable to conflate the multiple clustering structures that may arise from these distinct processes. To address this, we propose a multi-view Bayesian mixture model that identifies groups of variables (``views"), each of which defines a distinct clustering structure. We consider applications in stratified medicine, for which our principal goal is to identify clusters of patients that define distinct, clinically actionable disease subtypes. We adopt the semi-supervised, outcome-guided mixture modelling approach of Bayesian profile regression that makes use of a response variable in order to guide inference toward the clusterings that are most relevant in a stratified medicine context. We present the model, together with illustrative simulation examples, and examples from pan-cancer proteomics. We demonstrate how the approach can be used to perform integrative clustering, and consider an example in which different 'omics datasets are integrated in the context of breast cancer subtyping.
Audio-based AI classifiers show no evidence of improved COVID-19 screening over simple symptoms checkers
Dec 15, 2022



Abstract:Recent work has reported that AI classifiers trained on audio recordings can accurately predict severe acute respiratory syndrome coronavirus 2 (SARSCoV2) infection status. Here, we undertake a large scale study of audio-based deep learning classifiers, as part of the UK governments pandemic response. We collect and analyse a dataset of audio recordings from 67,842 individuals with linked metadata, including reverse transcription polymerase chain reaction (PCR) test outcomes, of whom 23,514 tested positive for SARS CoV 2. Subjects were recruited via the UK governments National Health Service Test-and-Trace programme and the REal-time Assessment of Community Transmission (REACT) randomised surveillance survey. In an unadjusted analysis of our dataset AI classifiers predict SARS-CoV-2 infection status with high accuracy (Receiver Operating Characteristic Area Under the Curve (ROCAUC) 0.846 [0.838, 0.854]) consistent with the findings of previous studies. However, after matching on measured confounders, such as age, gender, and self reported symptoms, our classifiers performance is much weaker (ROC-AUC 0.619 [0.594, 0.644]). Upon quantifying the utility of audio based classifiers in practical settings, we find them to be outperformed by simple predictive scores based on user reported symptoms.
A large-scale and PCR-referenced vocal audio dataset for COVID-19
Dec 15, 2022Abstract:The UK COVID-19 Vocal Audio Dataset is designed for the training and evaluation of machine learning models that classify SARS-CoV-2 infection status or associated respiratory symptoms using vocal audio. The UK Health Security Agency recruited voluntary participants through the national Test and Trace programme and the REACT-1 survey in England from March 2021 to March 2022, during dominant transmission of the Alpha and Delta SARS-CoV-2 variants and some Omicron variant sublineages. Audio recordings of volitional coughs, exhalations, and speech were collected in the 'Speak up to help beat coronavirus' digital survey alongside demographic, self-reported symptom and respiratory condition data, and linked to SARS-CoV-2 test results. The UK COVID-19 Vocal Audio Dataset represents the largest collection of SARS-CoV-2 PCR-referenced audio recordings to date. PCR results were linked to 70,794 of 72,999 participants and 24,155 of 25,776 positive cases. Respiratory symptoms were reported by 45.62% of participants. This dataset has additional potential uses for bioacoustics research, with 11.30% participants reporting asthma, and 27.20% with linked influenza PCR test results.
Kernel learning approaches for summarising and combining posterior similarity matrices
Sep 27, 2020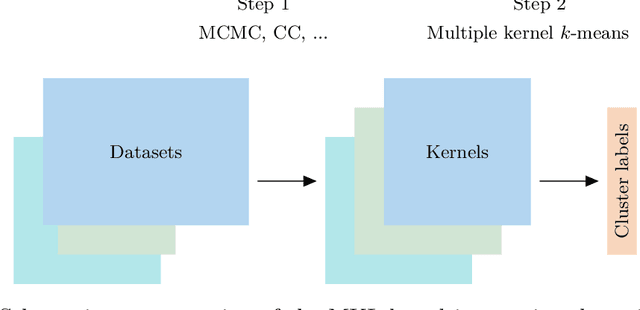

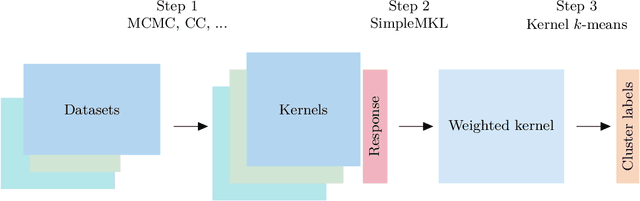
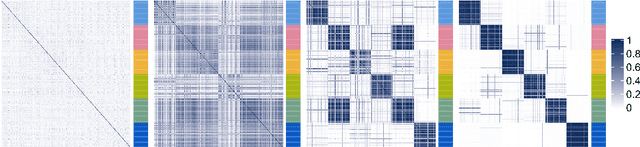
Abstract:When using Markov chain Monte Carlo (MCMC) algorithms to perform inference for Bayesian clustering models, such as mixture models, the output is typically a sample of clusterings (partitions) drawn from the posterior distribution. In practice, a key challenge is how to summarise this output. Here we build upon the notion of the posterior similarity matrix (PSM) in order to suggest new approaches for summarising the output of MCMC algorithms for Bayesian clustering models. A key contribution of our work is the observation that PSMs are positive semi-definite, and hence can be used to define probabilistically-motivated kernel matrices that capture the clustering structure present in the data. This observation enables us to employ a range of kernel methods to obtain summary clusterings, and otherwise exploit the information summarised by PSMs. For example, if we have multiple PSMs, each corresponding to a different dataset on a common set of statistical units, we may use standard methods for combining kernels in order to perform integrative clustering. We may moreover embed PSMs within predictive kernel models in order to perform outcome-guided data integration. We demonstrate the performances of the proposed methods through a range of simulation studies as well as two real data applications. R code is available at https://github.com/acabassi/combine-psms.
Distributed Bayesian Computation for Model Choice
Oct 10, 2019



Abstract:We propose a general method for distributed Bayesian model choice, where each worker has access only to non-overlapping subsets of the data. Our approach approximates the model evidence for the full data set through Monte Carlo sampling from the posterior on every subset generating a model evidence per subset. The model evidences per worker are then consistently combined using a novel approach which corrects for the splitting using summary statistics of the generated samples. This divide-and-conquer approach allows Bayesian model choice in the large data setting, exploiting all available information but limiting communication between workers. Our work thereby complements the work on consensus Monte Carlo (Scott et al., 2016) by explicitly enabling model choice. In addition, we show how the suggested approach can be extended to model choice within a reversible jump setting that explores multiple models within one run.
High-dimensional regression in practice: an empirical study of finite-sample prediction, variable selection and ranking
Aug 02, 2018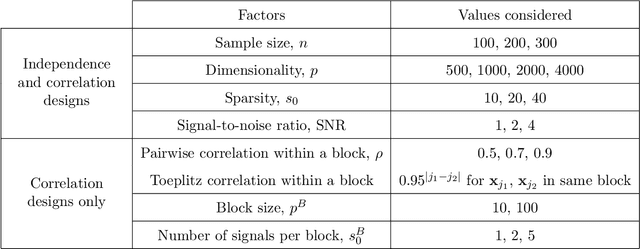
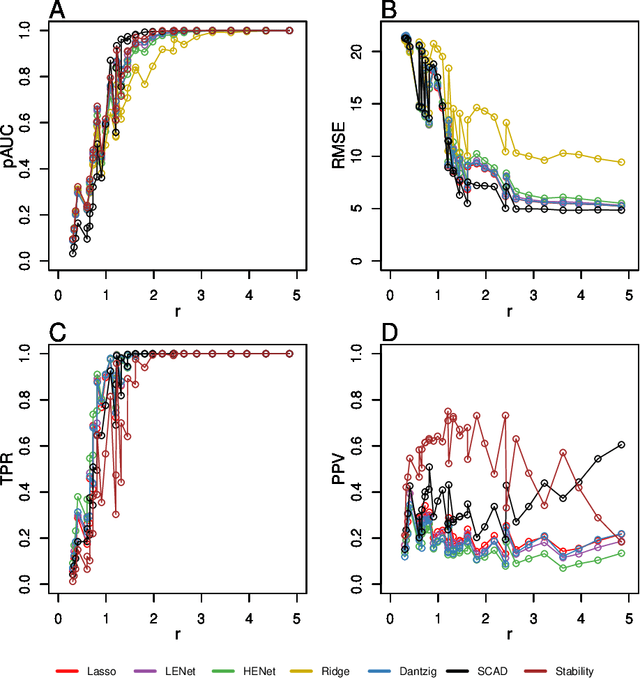
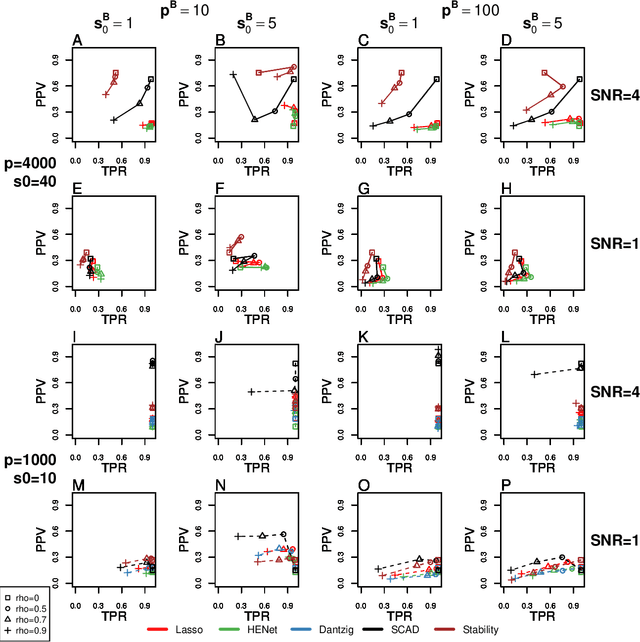
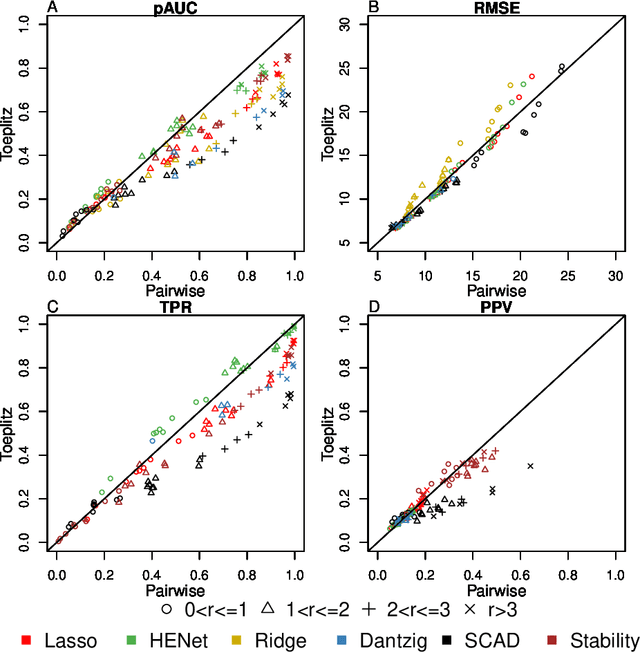
Abstract:Penalized likelihood methods are widely used for high-dimensional regression. Although many methods have been proposed and the associated theory is now well-developed, the relative efficacy of different methods in finite-sample settings, as encountered in practice, remains incompletely understood. There is therefore a need for empirical investigations in this area that can offer practical insight and guidance to users of these methods. In this paper we present a large-scale comparison of penalized regression methods. We distinguish between three related goals: prediction, variable selection and variable ranking. Our results span more than 1,800 data-generating scenarios, allowing us to systematically consider the influence of various factors (sample size, dimensionality, sparsity, signal strength and multicollinearity). We consider several widely-used methods (Lasso, Elastic Net, Ridge Regression, SCAD, the Dantzig Selector as well as Stability Selection). We find considerable variation in performance between methods, with results dependent on details of the data-generating scenario and the specific goal. Our results support a `no panacea' view, with no unambiguous winner across all scenarios, even in this restricted setting where all data align well with the assumptions underlying the methods. Lasso is well-behaved, performing competitively in many scenarios, while SCAD is highly variable. Substantial benefits from a Ridge-penalty are only seen in the most challenging scenarios with strong multi-collinearity. The results are supported by semi-synthetic analyzes using gene expression data from cancer samples. Our empirical results complement existing theory and provide a resource to compare methods across a range of scenarios and metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge