Ivan Kiskin
Few-shot bioacoustic event detection at the DCASE 2023 challenge
Jun 15, 2023



Abstract:Few-shot bioacoustic event detection consists in detecting sound events of specified types, in varying soundscapes, while having access to only a few examples of the class of interest. This task ran as part of the DCASE challenge for the third time this year with an evaluation set expanded to include new animal species, and a new rule: ensemble models were no longer allowed. The 2023 few shot task received submissions from 6 different teams with F-scores reaching as high as 63% on the evaluation set. Here we describe the task, focusing on describing the elements that differed from previous years. We also take a look back at past editions to describe how the task has evolved. Not only have the F-score results steadily improved (40% to 60% to 63%), but the type of systems proposed have also become more complex. Sound event detection systems are no longer simple variations of the baselines provided: multiple few-shot learning methodologies are still strong contenders for the task.
Dual Bayesian ResNet: A Deep Learning Approach to Heart Murmur Detection
May 26, 2023Abstract:This study presents our team PathToMyHeart's contribution to the George B. Moody PhysioNet Challenge 2022. Two models are implemented. The first model is a Dual Bayesian ResNet (DBRes), where each patient's recording is segmented into overlapping log mel spectrograms. These undergo two binary classifications: present versus unknown or absent, and unknown versus present or absent. The classifications are aggregated to give a patient's final classification. The second model is the output of DBRes integrated with demographic data and signal features using XGBoost.DBRes achieved our best weighted accuracy of $0.771$ on the hidden test set for murmur classification, which placed us fourth for the murmur task. (On the clinical outcome task, which we neglected, we scored 17th with costs of $12637$.) On our held-out subset of the training set, integrating the demographic data and signal features improved DBRes's accuracy from $0.762$ to $0.820$. However, this decreased DBRes's weighted accuracy from $0.780$ to $0.749$. Our results demonstrate that log mel spectrograms are an effective representation of heart sound recordings, Bayesian networks provide strong supervised classification performance, and treating the ternary classification as two binary classifications increases performance on the weighted accuracy.
* 5 pages, 3 figures
Learning to detect an animal sound from five examples
May 22, 2023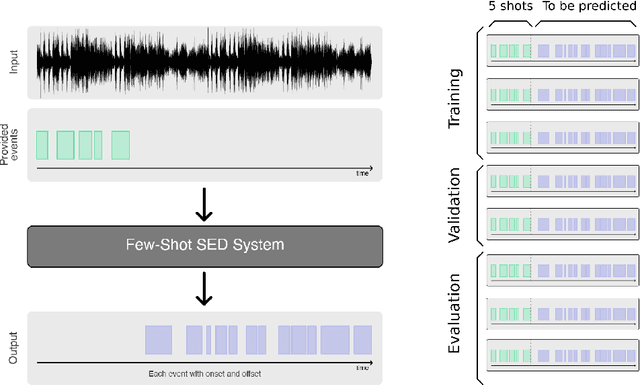
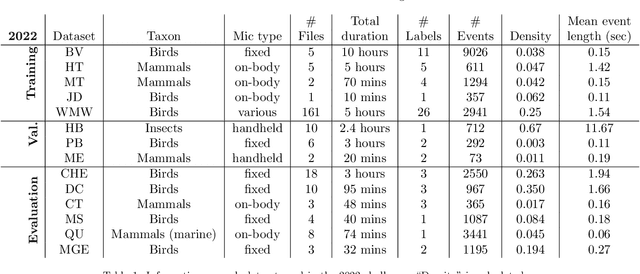
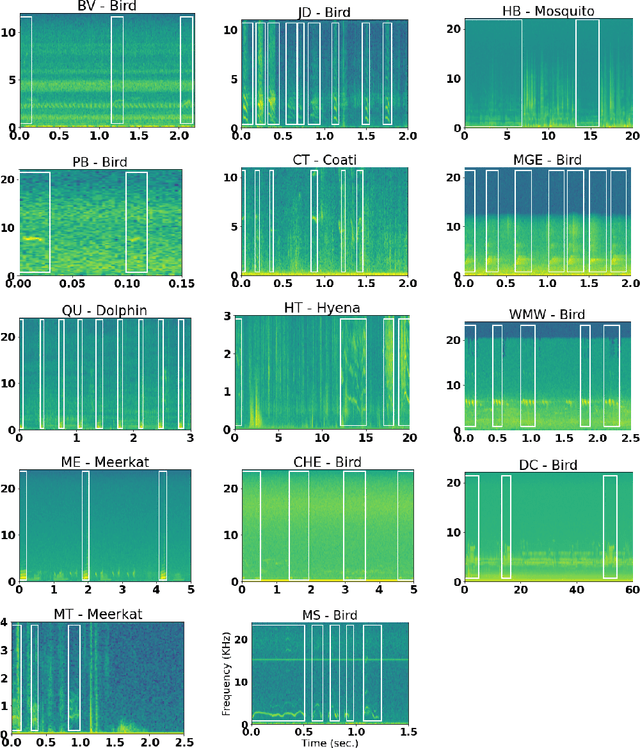
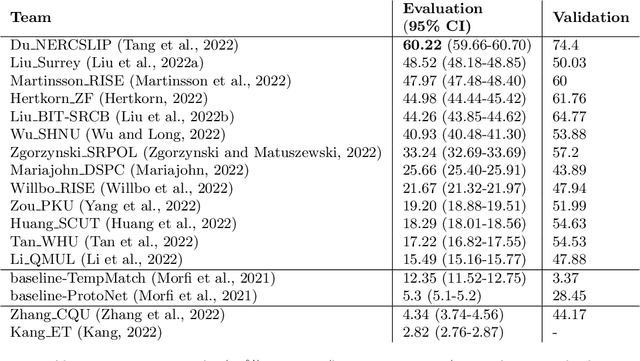
Abstract:Automatic detection and classification of animal sounds has many applications in biodiversity monitoring and animal behaviour. In the past twenty years, the volume of digitised wildlife sound available has massively increased, and automatic classification through deep learning now shows strong results. However, bioacoustics is not a single task but a vast range of small-scale tasks (such as individual ID, call type, emotional indication) with wide variety in data characteristics, and most bioacoustic tasks do not come with strongly-labelled training data. The standard paradigm of supervised learning, focussed on a single large-scale dataset and/or a generic pre-trained algorithm, is insufficient. In this work we recast bioacoustic sound event detection within the AI framework of few-shot learning. We adapt this framework to sound event detection, such that a system can be given the annotated start/end times of as few as 5 events, and can then detect events in long-duration audio -- even when the sound category was not known at the time of algorithm training. We introduce a collection of open datasets designed to strongly test a system's ability to perform few-shot sound event detections, and we present the results of a public contest to address the task. We show that prototypical networks are a strong-performing method, when enhanced with adaptations for general characteristics of animal sounds. We demonstrate that widely-varying sound event durations are an important factor in performance, as well as non-stationarity, i.e. gradual changes in conditions throughout the duration of a recording. For fine-grained bioacoustic recognition tasks without massive annotated training data, our results demonstrate that few-shot sound event detection is a powerful new method, strongly outperforming traditional signal-processing detection methods in the fully automated scenario.
Audio-based AI classifiers show no evidence of improved COVID-19 screening over simple symptoms checkers
Dec 15, 2022



Abstract:Recent work has reported that AI classifiers trained on audio recordings can accurately predict severe acute respiratory syndrome coronavirus 2 (SARSCoV2) infection status. Here, we undertake a large scale study of audio-based deep learning classifiers, as part of the UK governments pandemic response. We collect and analyse a dataset of audio recordings from 67,842 individuals with linked metadata, including reverse transcription polymerase chain reaction (PCR) test outcomes, of whom 23,514 tested positive for SARS CoV 2. Subjects were recruited via the UK governments National Health Service Test-and-Trace programme and the REal-time Assessment of Community Transmission (REACT) randomised surveillance survey. In an unadjusted analysis of our dataset AI classifiers predict SARS-CoV-2 infection status with high accuracy (Receiver Operating Characteristic Area Under the Curve (ROCAUC) 0.846 [0.838, 0.854]) consistent with the findings of previous studies. However, after matching on measured confounders, such as age, gender, and self reported symptoms, our classifiers performance is much weaker (ROC-AUC 0.619 [0.594, 0.644]). Upon quantifying the utility of audio based classifiers in practical settings, we find them to be outperformed by simple predictive scores based on user reported symptoms.
A large-scale and PCR-referenced vocal audio dataset for COVID-19
Dec 15, 2022Abstract:The UK COVID-19 Vocal Audio Dataset is designed for the training and evaluation of machine learning models that classify SARS-CoV-2 infection status or associated respiratory symptoms using vocal audio. The UK Health Security Agency recruited voluntary participants through the national Test and Trace programme and the REACT-1 survey in England from March 2021 to March 2022, during dominant transmission of the Alpha and Delta SARS-CoV-2 variants and some Omicron variant sublineages. Audio recordings of volitional coughs, exhalations, and speech were collected in the 'Speak up to help beat coronavirus' digital survey alongside demographic, self-reported symptom and respiratory condition data, and linked to SARS-CoV-2 test results. The UK COVID-19 Vocal Audio Dataset represents the largest collection of SARS-CoV-2 PCR-referenced audio recordings to date. PCR results were linked to 70,794 of 72,999 participants and 24,155 of 25,776 positive cases. Respiratory symptoms were reported by 45.62% of participants. This dataset has additional potential uses for bioacoustics research, with 11.30% participants reporting asthma, and 27.20% with linked influenza PCR test results.
Statistical Design and Analysis for Robust Machine Learning: A Case Study from COVID-19
Dec 15, 2022



Abstract:Since early in the coronavirus disease 2019 (COVID-19) pandemic, there has been interest in using artificial intelligence methods to predict COVID-19 infection status based on vocal audio signals, for example cough recordings. However, existing studies have limitations in terms of data collection and of the assessment of the performances of the proposed predictive models. This paper rigorously assesses state-of-the-art machine learning techniques used to predict COVID-19 infection status based on vocal audio signals, using a dataset collected by the UK Health Security Agency. This dataset includes acoustic recordings and extensive study participant meta-data. We provide guidelines on testing the performance of methods to classify COVID-19 infection status based on acoustic features and we discuss how these can be extended more generally to the development and assessment of predictive methods based on public health datasets.
The ACM Multimedia 2022 Computational Paralinguistics Challenge: Vocalisations, Stuttering, Activity, & Mosquitoes
May 13, 2022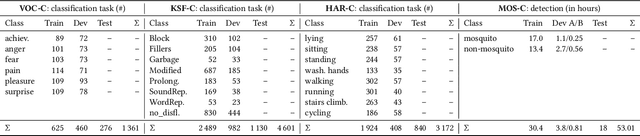
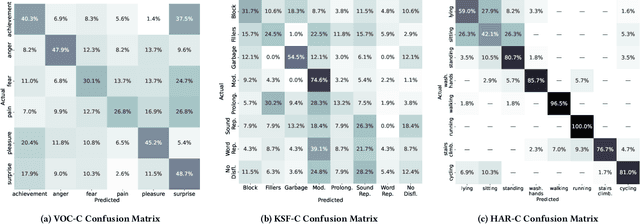
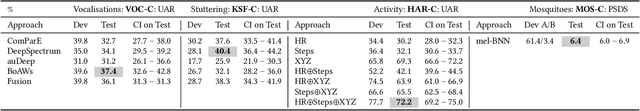
Abstract:The ACM Multimedia 2022 Computational Paralinguistics Challenge addresses four different problems for the first time in a research competition under well-defined conditions: In the Vocalisations and Stuttering Sub-Challenges, a classification on human non-verbal vocalisations and speech has to be made; the Activity Sub-Challenge aims at beyond-audio human activity recognition from smartwatch sensor data; and in the Mosquitoes Sub-Challenge, mosquitoes need to be detected. We describe the Sub-Challenges, baseline feature extraction, and classifiers based on the usual ComPaRE and BoAW features, the auDeep toolkit, and deep feature extraction from pre-trained CNNs using the DeepSpectRum toolkit; in addition, we add end-to-end sequential modelling, and a log-mel-128-BNN.
HumBugDB: A Large-scale Acoustic Mosquito Dataset
Oct 14, 2021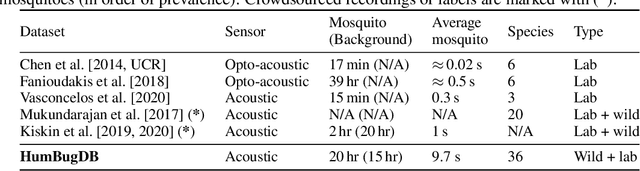
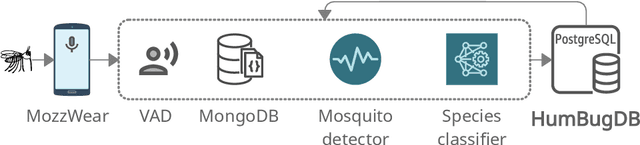
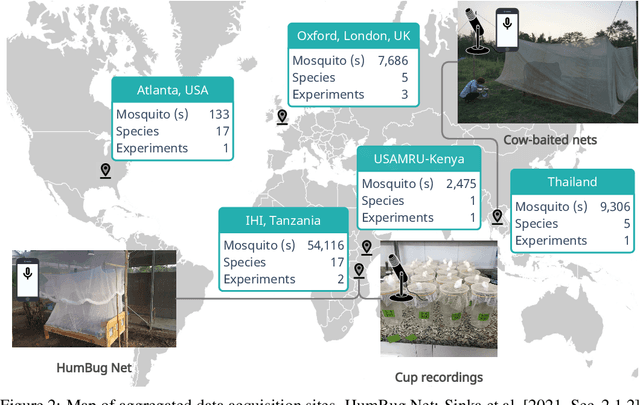
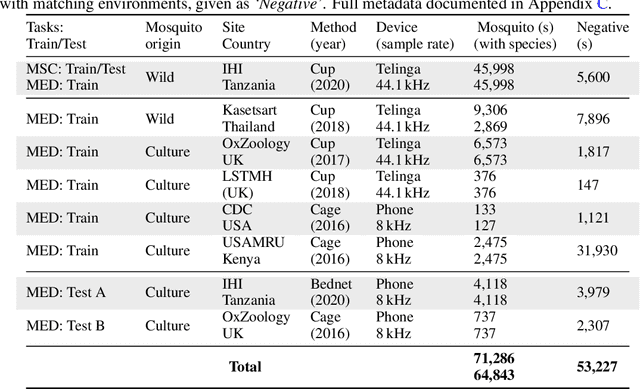
Abstract:This paper presents the first large-scale multi-species dataset of acoustic recordings of mosquitoes tracked continuously in free flight. We present 20 hours of audio recordings that we have expertly labelled and tagged precisely in time. Significantly, 18 hours of recordings contain annotations from 36 different species. Mosquitoes are well-known carriers of diseases such as malaria, dengue and yellow fever. Collecting this dataset is motivated by the need to assist applications which utilise mosquito acoustics to conduct surveys to help predict outbreaks and inform intervention policy. The task of detecting mosquitoes from the sound of their wingbeats is challenging due to the difficulty in collecting recordings from realistic scenarios. To address this, as part of the HumBug project, we conducted global experiments to record mosquitoes ranging from those bred in culture cages to mosquitoes captured in the wild. Consequently, the audio recordings vary in signal-to-noise ratio and contain a broad range of indoor and outdoor background environments from Tanzania, Thailand, Kenya, the USA and the UK. In this paper we describe in detail how we collected, labelled and curated the data. The data is provided from a PostgreSQL database, which contains important metadata such as the capture method, age, feeding status and gender of the mosquitoes. Additionally, we provide code to extract features and train Bayesian convolutional neural networks for two key tasks: the identification of mosquitoes from their corresponding background environments, and the classification of detected mosquitoes into species. Our extensive dataset is both challenging to machine learning researchers focusing on acoustic identification, and critical to entomologists, geo-spatial modellers and other domain experts to understand mosquito behaviour, model their distribution, and manage the threat they pose to humans.
HumBug Zooniverse: a crowd-sourced acoustic mosquito dataset
Feb 14, 2020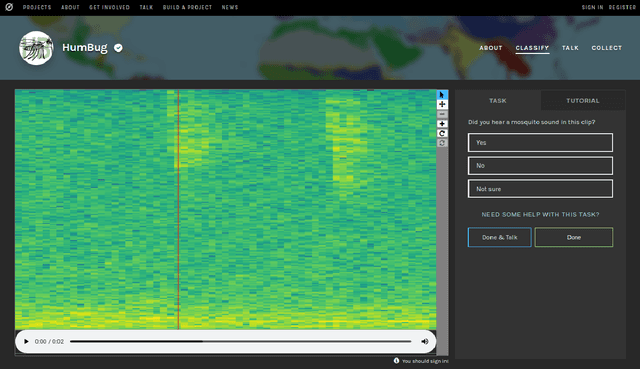
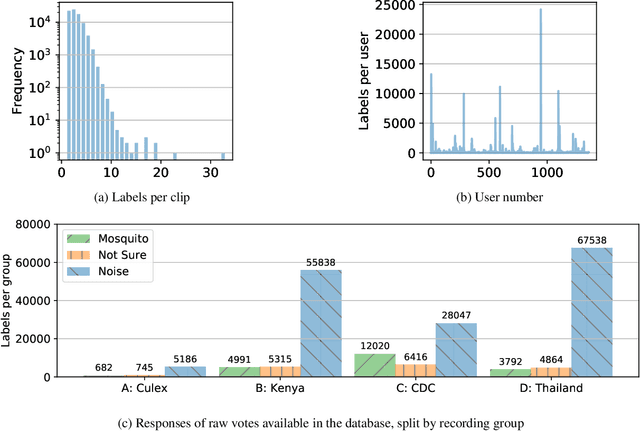
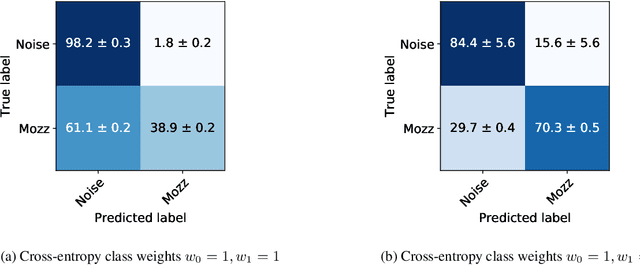
Abstract:Mosquitoes are the only known vector of malaria, which leads to hundreds of thousands of deaths each year. Understanding the number and location of potential mosquito vectors is of paramount importance to aid the reduction of malaria transmission cases. In recent years, deep learning has become widely used for bioacoustic classification tasks. In order to enable further research applications in this field, we release a new dataset of mosquito audio recordings. With over a thousand contributors, we obtained 195,434 labels of two second duration, of which approximately 10 percent signify mosquito events. We present an example use of the dataset, in which we train a convolutional neural network on log-Mel features, showcasing the information content of the labels. We hope this will become a vital resource for those researching all aspects of malaria, and add to the existing audio datasets for bioacoustic detection and signal processing.
Super-resolution of Time-series Labels for Bootstrapped Event Detection
Jun 01, 2019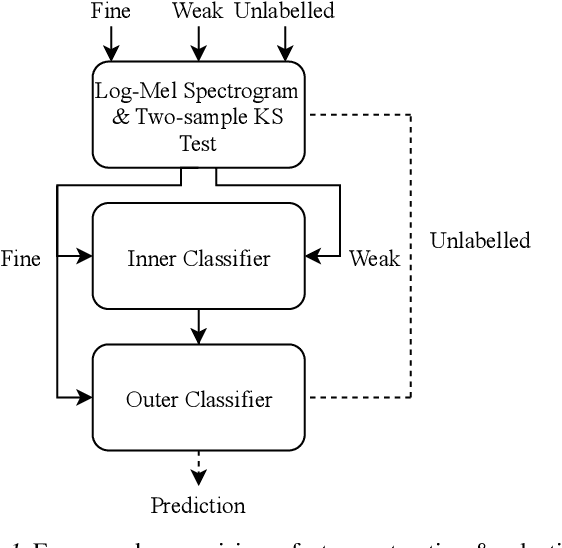
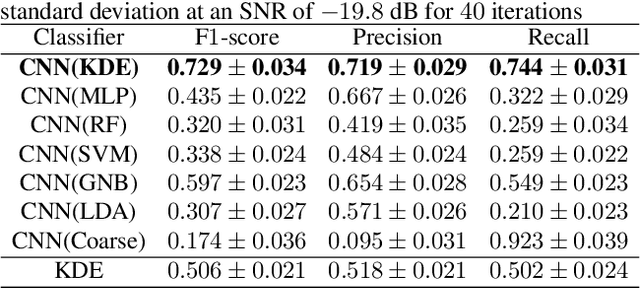
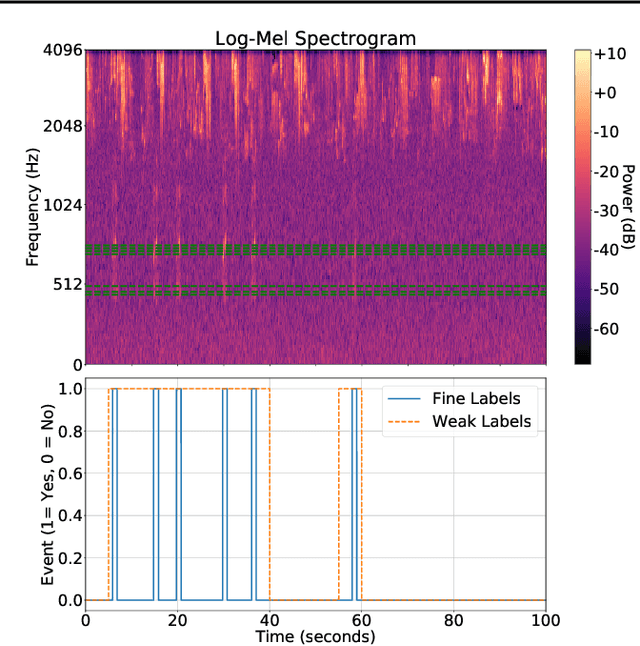
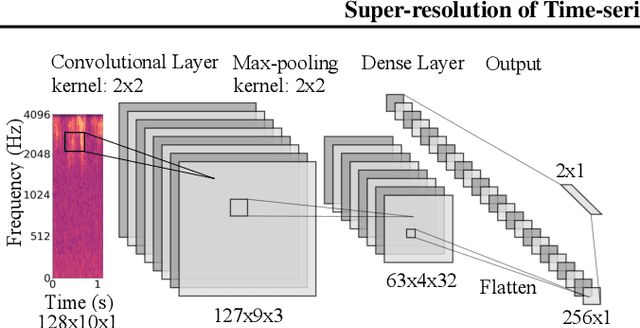
Abstract:Solving real-world problems, particularly with deep learning, relies on the availability of abundant, quality data. In this paper we develop a novel framework that maximises the utility of time-series datasets that contain only small quantities of expertly-labelled data, larger quantities of weakly (or coarsely) labelled data and a large volume of unlabelled data. This represents scenarios commonly encountered in the real world, such as in crowd-sourcing applications. In our work, we use a nested loop using a Kernel Density Estimator (KDE) to super-resolve the abundant low-quality data labels, thereby enabling effective training of a Convolutional Neural Network (CNN). We demonstrate two key results: a) The KDE is able to super-resolve labels more accurately, and with better calibrated probabilities, than well-established classifiers acting as baselines; b) Our CNN, trained on super-resolved labels from the KDE, achieves an improvement in F1 score of 22.1% over the next best baseline system in our candidate problem domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge