Guy Parsons
Clinical knowledge in LLMs does not translate to human interactions
Apr 26, 2025Abstract:Global healthcare providers are exploring use of large language models (LLMs) to provide medical advice to the public. LLMs now achieve nearly perfect scores on medical licensing exams, but this does not necessarily translate to accurate performance in real-world settings. We tested if LLMs can assist members of the public in identifying underlying conditions and choosing a course of action (disposition) in ten medical scenarios in a controlled study with 1,298 participants. Participants were randomly assigned to receive assistance from an LLM (GPT-4o, Llama 3, Command R+) or a source of their choice (control). Tested alone, LLMs complete the scenarios accurately, correctly identifying conditions in 94.9% of cases and disposition in 56.3% on average. However, participants using the same LLMs identified relevant conditions in less than 34.5% of cases and disposition in less than 44.2%, both no better than the control group. We identify user interactions as a challenge to the deployment of LLMs for medical advice. Standard benchmarks for medical knowledge and simulated patient interactions do not predict the failures we find with human participants. Moving forward, we recommend systematic human user testing to evaluate interactive capabilities prior to public deployments in healthcare.
Multimodal deep learning approach to predicting neurological recovery from coma after cardiac arrest
Mar 09, 2024



Abstract:This work showcases our team's (The BEEGees) contributions to the 2023 George B. Moody PhysioNet Challenge. The aim was to predict neurological recovery from coma following cardiac arrest using clinical data and time-series such as multi-channel EEG and ECG signals. Our modelling approach is multimodal, based on two-dimensional spectrogram representations derived from numerous EEG channels, alongside the integration of clinical data and features extracted directly from EEG recordings. Our submitted model achieved a Challenge score of $0.53$ on the hidden test set for predictions made $72$ hours after return of spontaneous circulation. Our study shows the efficacy and limitations of employing transfer learning in medical classification. With regard to prospective implementation, our analysis reveals that the performance of the model is strongly linked to the selection of a decision threshold and exhibits strong variability across data splits.
Unsupervised Learning Approaches for Identifying ICU Patient Subgroups: Do Results Generalise?
Mar 05, 2024Abstract:The use of unsupervised learning to identify patient subgroups has emerged as a potentially promising direction to improve the efficiency of Intensive Care Units (ICUs). By identifying subgroups of patients with similar levels of medical resource need, ICUs could be restructured into a collection of smaller subunits, each catering to a specific group. However, it is unclear whether common patient subgroups exist across different ICUs, which would determine whether ICU restructuring could be operationalised in a standardised manner. In this paper, we tested the hypothesis that common ICU patient subgroups exist by examining whether the results from one existing study generalise to a different dataset. We extracted 16 features representing medical resource need and used consensus clustering to derive patient subgroups, replicating the previous study. We found limited similarities between our results and those of the previous study, providing evidence against the hypothesis. Our findings imply that there is significant variation between ICUs; thus, a standardised restructuring approach is unlikely to be appropriate. Instead, potential efficiency gains might be greater when the number and nature of the subunits are tailored to each ICU individually.
Review of multimodal machine learning approaches in healthcare
Feb 12, 2024Abstract:Machine learning methods in healthcare have traditionally focused on using data from a single modality, limiting their ability to effectively replicate the clinical practice of integrating multiple sources of information for improved decision making. Clinicians typically rely on a variety of data sources including patients' demographic information, laboratory data, vital signs and various imaging data modalities to make informed decisions and contextualise their findings. Recent advances in machine learning have facilitated the more efficient incorporation of multimodal data, resulting in applications that better represent the clinician's approach. Here, we provide a review of multimodal machine learning approaches in healthcare, offering a comprehensive overview of recent literature. We discuss the various data modalities used in clinical diagnosis, with a particular emphasis on imaging data. We evaluate fusion techniques, explore existing multimodal datasets and examine common training strategies.
Dual Bayesian ResNet: A Deep Learning Approach to Heart Murmur Detection
May 26, 2023Abstract:This study presents our team PathToMyHeart's contribution to the George B. Moody PhysioNet Challenge 2022. Two models are implemented. The first model is a Dual Bayesian ResNet (DBRes), where each patient's recording is segmented into overlapping log mel spectrograms. These undergo two binary classifications: present versus unknown or absent, and unknown versus present or absent. The classifications are aggregated to give a patient's final classification. The second model is the output of DBRes integrated with demographic data and signal features using XGBoost.DBRes achieved our best weighted accuracy of $0.771$ on the hidden test set for murmur classification, which placed us fourth for the murmur task. (On the clinical outcome task, which we neglected, we scored 17th with costs of $12637$.) On our held-out subset of the training set, integrating the demographic data and signal features improved DBRes's accuracy from $0.762$ to $0.820$. However, this decreased DBRes's weighted accuracy from $0.780$ to $0.749$. Our results demonstrate that log mel spectrograms are an effective representation of heart sound recordings, Bayesian networks provide strong supervised classification performance, and treating the ternary classification as two binary classifications increases performance on the weighted accuracy.
* 5 pages, 3 figures
Explaining Chest X-ray Pathologies in Natural Language
Jul 09, 2022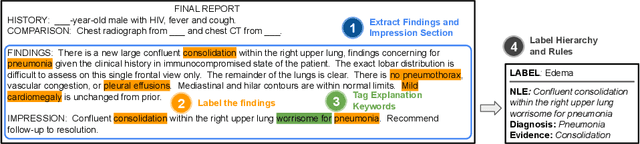
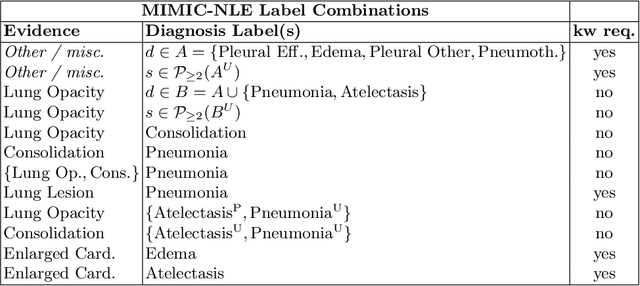
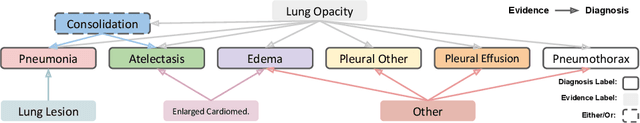
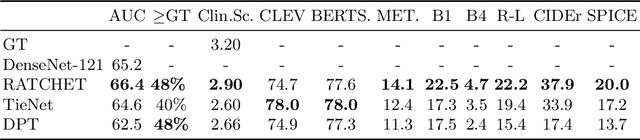
Abstract:Most deep learning algorithms lack explanations for their predictions, which limits their deployment in clinical practice. Approaches to improve explainability, especially in medical imaging, have often been shown to convey limited information, be overly reassuring, or lack robustness. In this work, we introduce the task of generating natural language explanations (NLEs) to justify predictions made on medical images. NLEs are human-friendly and comprehensive, and enable the training of intrinsically explainable models. To this goal, we introduce MIMIC-NLE, the first, large-scale, medical imaging dataset with NLEs. It contains over 38,000 NLEs, which explain the presence of various thoracic pathologies and chest X-ray findings. We propose a general approach to solve the task and evaluate several architectures on this dataset, including via clinician assessment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge