Maxime Kayser
M.D.
brat: Aligned Multi-View Embeddings for Brain MRI Analysis
Dec 21, 2025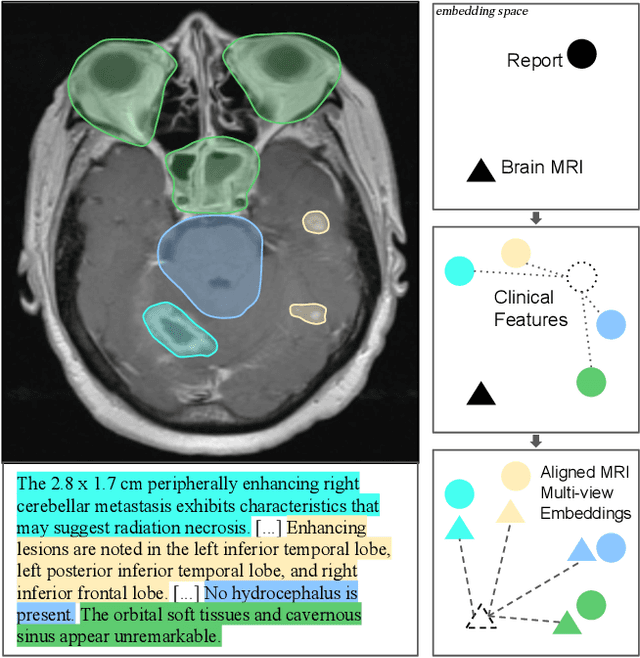
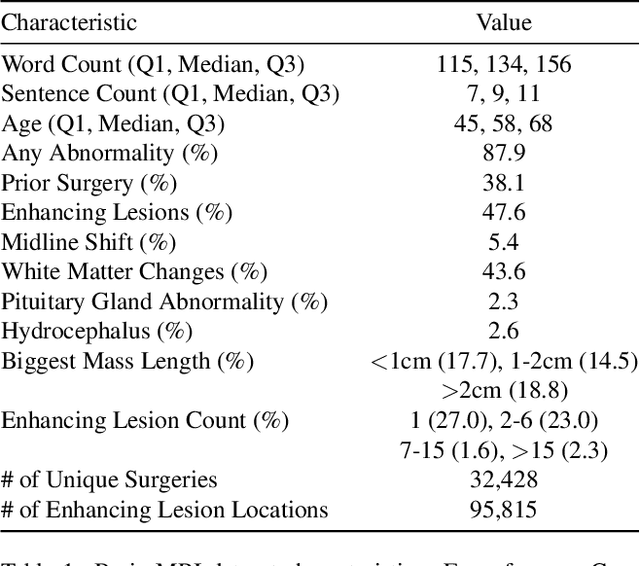
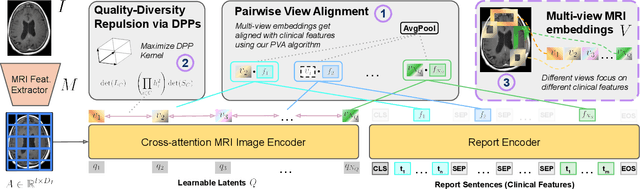
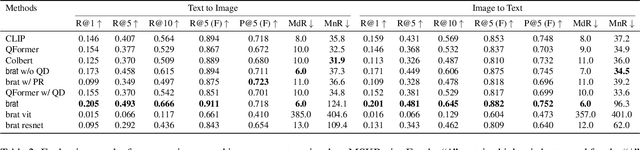
Abstract:We present brat (brain report alignment transformer), a multi-view representation learning framework for brain magnetic resonance imaging (MRI) trained on MRIs paired with clinical reports. Brain MRIs present unique challenges due to the presence of numerous, highly varied, and often subtle abnormalities that are localized to a few slices within a 3D volume. To address these challenges, we introduce a brain MRI dataset $10\times$ larger than existing ones, containing approximately 80,000 3D scans with corresponding radiology reports, and propose a multi-view pre-training approach inspired by advances in document retrieval. We develop an implicit query-feature matching mechanism and adopt concepts from quality-diversity to obtain multi-view embeddings of MRIs that are aligned with the clinical features given by report sentences. We evaluate our approach across multiple vision-language and vision tasks, demonstrating substantial performance improvements. The brat foundation models are publicly released.
Shh, don't say that! Domain Certification in LLMs
Feb 26, 2025Abstract:Large language models (LLMs) are often deployed to perform constrained tasks, with narrow domains. For example, customer support bots can be built on top of LLMs, relying on their broad language understanding and capabilities to enhance performance. However, these LLMs are adversarially susceptible, potentially generating outputs outside the intended domain. To formalize, assess, and mitigate this risk, we introduce domain certification; a guarantee that accurately characterizes the out-of-domain behavior of language models. We then propose a simple yet effective approach, which we call VALID that provides adversarial bounds as a certificate. Finally, we evaluate our method across a diverse set of datasets, demonstrating that it yields meaningful certificates, which bound the probability of out-of-domain samples tightly with minimum penalty to refusal behavior.
* 10 pages, includes appendix Published in International Conference on Learning Representations (ICLR) 2025
Fool Me Once? Contrasting Textual and Visual Explanations in a Clinical Decision-Support Setting
Oct 16, 2024



Abstract:The growing capabilities of AI models are leading to their wider use, including in safety-critical domains. Explainable AI (XAI) aims to make these models safer to use by making their inference process more transparent. However, current explainability methods are seldom evaluated in the way they are intended to be used: by real-world end users. To address this, we conducted a large-scale user study with 85 healthcare practitioners in the context of human-AI collaborative chest X-ray analysis. We evaluated three types of explanations: visual explanations (saliency maps), natural language explanations, and a combination of both modalities. We specifically examined how different explanation types influence users depending on whether the AI advice and explanations are factually correct. We find that text-based explanations lead to significant over-reliance, which is alleviated by combining them with saliency maps. We also observe that the quality of explanations, that is, how much factually correct information they entail, and how much this aligns with AI correctness, significantly impacts the usefulness of the different explanation types.
Explaining Chest X-ray Pathologies in Natural Language
Jul 09, 2022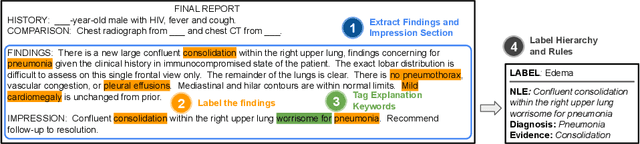
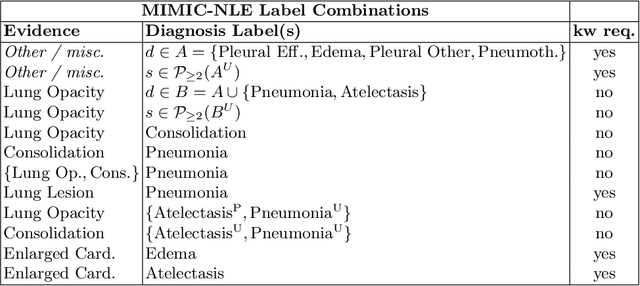
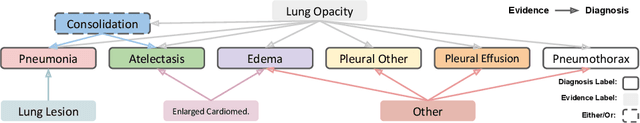
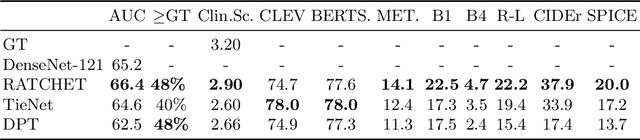
Abstract:Most deep learning algorithms lack explanations for their predictions, which limits their deployment in clinical practice. Approaches to improve explainability, especially in medical imaging, have often been shown to convey limited information, be overly reassuring, or lack robustness. In this work, we introduce the task of generating natural language explanations (NLEs) to justify predictions made on medical images. NLEs are human-friendly and comprehensive, and enable the training of intrinsically explainable models. To this goal, we introduce MIMIC-NLE, the first, large-scale, medical imaging dataset with NLEs. It contains over 38,000 NLEs, which explain the presence of various thoracic pathologies and chest X-ray findings. We propose a general approach to solve the task and evaluate several architectures on this dataset, including via clinician assessment.
e-ViL: A Dataset and Benchmark for Natural Language Explanations in Vision-Language Tasks
May 08, 2021


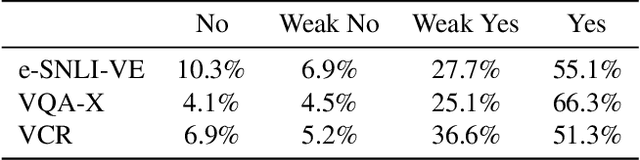
Abstract:Recently, an increasing number of works have introduced models capable of generating natural language explanations (NLEs) for their predictions on vision-language (VL) tasks. Such models are appealing because they can provide human-friendly and comprehensive explanations. However, there is still a lack of unified evaluation approaches for the explanations generated by these models. Moreover, there are currently only few datasets of NLEs for VL tasks. In this work, we introduce e-ViL, a benchmark for explainable vision-language tasks that establishes a unified evaluation framework and provides the first comprehensive comparison of existing approaches that generate NLEs for VL tasks. e-ViL spans four models and three datasets. Both automatic metrics and human evaluation are used to assess model-generated explanations. We also introduce e-SNLI-VE, the largest existing VL dataset with NLEs (over 430k instances). Finally, we propose a new model that combines UNITER, which learns joint embeddings of images and text, and GPT-2, a pre-trained language model that is well-suited for text generation. It surpasses the previous state-of-the-art by a large margin across all datasets.
A learning without forgetting approach to incorporate artifact knowledge in polyp localization tasks
Feb 11, 2020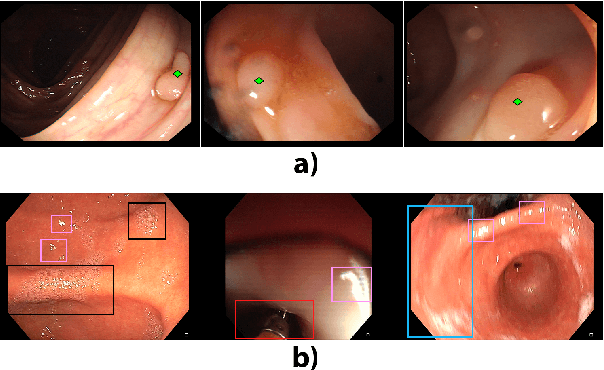
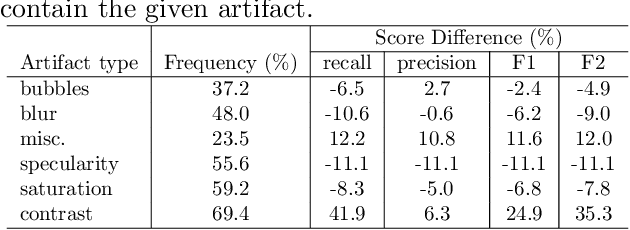
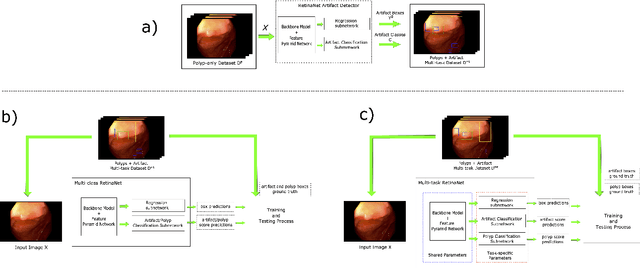
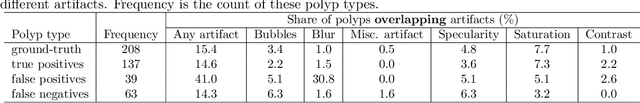
Abstract:Colorectal polyps are abnormalities in the colon tissue that can develop into colorectal cancer. The survival rate for patients is higher when the disease is detected at an early stage and polyps can be removed before they develop into malignant tumors. Deep learning methods have become the state of art in automatic polyp detection. However, the performance of current models heavily relies on the size and quality of the training datasets. Endoscopic video sequences tend to be corrupted by different artifacts affecting visibility and hence, the detection rates. In this work, we analyze the effects that artifacts have in the polyp localization problem. For this, we evaluate the RetinaNet architecture, originally defined for object localization. We also define a model inspired by the learning without forgetting framework, which allows us to employ artifact detection knowledge in the polyp localization problem. Finally, we perform several experiments to analyze the influence of the artifacts in the performance of these models. To our best knowledge, this is the first extensive analysis of the influence of artifact in polyp localization and the first work incorporating learning without forgetting ideas for simultaneous artifact and polyp localization tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge