Roger D. Soberanis-Mukul
M.D.
BronchOpt : Vision-Based Pose Optimization with Fine-Tuned Foundation Models for Accurate Bronchoscopy Navigation
Nov 12, 2025



Abstract:Accurate intra-operative localization of the bronchoscope tip relative to patient anatomy remains challenging due to respiratory motion, anatomical variability, and CT-to-body divergence that cause deformation and misalignment between intra-operative views and pre-operative CT. Existing vision-based methods often fail to generalize across domains and patients, leading to residual alignment errors. This work establishes a generalizable foundation for bronchoscopy navigation through a robust vision-based framework and a new synthetic benchmark dataset that enables standardized and reproducible evaluation. We propose a vision-based pose optimization framework for frame-wise 2D-3D registration between intra-operative endoscopic views and pre-operative CT anatomy. A fine-tuned modality- and domain-invariant encoder enables direct similarity computation between real endoscopic RGB frames and CT-rendered depth maps, while a differentiable rendering module iteratively refines camera poses through depth consistency. To enhance reproducibility, we introduce the first public synthetic benchmark dataset for bronchoscopy navigation, addressing the lack of paired CT-endoscopy data. Trained exclusively on synthetic data distinct from the benchmark, our model achieves an average translational error of 2.65 mm and a rotational error of 0.19 rad, demonstrating accurate and stable localization. Qualitative results on real patient data further confirm strong cross-domain generalization, achieving consistent frame-wise 2D-3D alignment without domain-specific adaptation. Overall, the proposed framework achieves robust, domain-invariant localization through iterative vision-based optimization, while the new benchmark provides a foundation for standardized progress in vision-based bronchoscopy navigation.
TwinOR: Photorealistic Digital Twins of Dynamic Operating Rooms for Embodied AI Research
Nov 10, 2025Abstract:Developing embodied AI for intelligent surgical systems requires safe, controllable environments for continual learning and evaluation. However, safety regulations and operational constraints in operating rooms (ORs) limit embodied agents from freely perceiving and interacting in realistic settings. Digital twins provide high-fidelity, risk-free environments for exploration and training. How we may create photorealistic and dynamic digital representations of ORs that capture relevant spatial, visual, and behavioral complexity remains unclear. We introduce TwinOR, a framework for constructing photorealistic, dynamic digital twins of ORs for embodied AI research. The system reconstructs static geometry from pre-scan videos and continuously models human and equipment motion through multi-view perception of OR activities. The static and dynamic components are fused into an immersive 3D environment that supports controllable simulation and embodied exploration. The proposed framework reconstructs complete OR geometry with centimeter level accuracy while preserving dynamic interaction across surgical workflows, enabling realistic renderings and a virtual playground for embodied AI systems. In our experiments, TwinOR simulates stereo and monocular sensor streams for geometry understanding and visual localization tasks. Models such as FoundationStereo and ORB-SLAM3 on TwinOR-synthesized data achieve performance within their reported accuracy on real indoor datasets, demonstrating that TwinOR provides sensor-level realism sufficient for perception and localization challenges. By establishing a real-to-sim pipeline for constructing dynamic, photorealistic digital twins of OR environments, TwinOR enables the safe, scalable, and data-efficient development and benchmarking of embodied AI, ultimately accelerating the deployment of embodied AI from sim-to-real.
Did you just see that? Arbitrary view synthesis for egocentric replay of operating room workflows from ambient sensors
Oct 06, 2025Abstract:Observing surgical practice has historically relied on fixed vantage points or recollections, leaving the egocentric visual perspectives that guide clinical decisions undocumented. Fixed-camera video can capture surgical workflows at the room-scale, but cannot reconstruct what each team member actually saw. Thus, these videos only provide limited insights into how decisions that affect surgical safety, training, and workflow optimization are made. Here we introduce EgoSurg, the first framework to reconstruct the dynamic, egocentric replays for any operating room (OR) staff directly from wall-mounted fixed-camera video, and thus, without intervention to clinical workflow. EgoSurg couples geometry-driven neural rendering with diffusion-based view enhancement, enabling high-visual fidelity synthesis of arbitrary and egocentric viewpoints at any moment. In evaluation across multi-site surgical cases and controlled studies, EgoSurg reconstructs person-specific visual fields and arbitrary viewpoints with high visual quality and fidelity. By transforming existing OR camera infrastructure into a navigable dynamic 3D record, EgoSurg establishes a new foundation for immersive surgical data science, enabling surgical practice to be visualized, experienced, and analyzed from every angle.
From Generalization to Precision: Exploring SAM for Tool Segmentation in Surgical Environments
Feb 28, 2024Abstract:Purpose: Accurate tool segmentation is essential in computer-aided procedures. However, this task conveys challenges due to artifacts' presence and the limited training data in medical scenarios. Methods that generalize to unseen data represent an interesting venue, where zero-shot segmentation presents an option to account for data limitation. Initial exploratory works with the Segment Anything Model (SAM) show that bounding-box-based prompting presents notable zero-short generalization. However, point-based prompting leads to a degraded performance that further deteriorates under image corruption. We argue that SAM drastically over-segment images with high corruption levels, resulting in degraded performance when only a single segmentation mask is considered, while the combination of the masks overlapping the object of interest generates an accurate prediction. Method: We use SAM to generate the over-segmented prediction of endoscopic frames. Then, we employ the ground-truth tool mask to analyze the results of SAM when the best single mask is selected as prediction and when all the individual masks overlapping the object of interest are combined to obtain the final predicted mask. We analyze the Endovis18 and Endovis17 instrument segmentation datasets using synthetic corruptions of various strengths and an In-House dataset featuring counterfactually created real-world corruptions. Results: Combining the over-segmented masks contributes to improvements in the IoU. Furthermore, selecting the best single segmentation presents a competitive IoU score for clean images. Conclusions: Combined SAM predictions present improved results and robustness up to a certain corruption level. However, appropriate prompting strategies are fundamental for implementing these models in the medical domain.
An Endoscopic Chisel: Intraoperative Imaging Carves 3D Anatomical Models
Feb 19, 2024Abstract:Purpose: Preoperative imaging plays a pivotal role in sinus surgery where CTs offer patient-specific insights of complex anatomy, enabling real-time intraoperative navigation to complement endoscopy imaging. However, surgery elicits anatomical changes not represented in the preoperative model, generating an inaccurate basis for navigation during surgery progression. Methods: We propose a first vision-based approach to update the preoperative 3D anatomical model leveraging intraoperative endoscopic video for navigated sinus surgery where relative camera poses are known. We rely on comparisons of intraoperative monocular depth estimates and preoperative depth renders to identify modified regions. The new depths are integrated in these regions through volumetric fusion in a truncated signed distance function representation to generate an intraoperative 3D model that reflects tissue manipulation. Results: We quantitatively evaluate our approach by sequentially updating models for a five-step surgical progression in an ex vivo specimen. We compute the error between correspondences from the updated model and ground-truth intraoperative CT in the region of anatomical modification. The resulting models show a decrease in error during surgical progression as opposed to increasing when no update is employed. Conclusion: Our findings suggest that preoperative 3D anatomical models can be updated using intraoperative endoscopy video in navigated sinus surgery. Future work will investigate improvements to monocular depth estimation as well as removing the need for external navigation systems. The resulting ability to continuously update the patient model may provide surgeons with a more precise understanding of the current anatomical state and paves the way toward a digital twin paradigm for sinus surgery.
A Quantitative Evaluation of Dense 3D Reconstruction of Sinus Anatomy from Monocular Endoscopic Video
Oct 22, 2023



Abstract:Generating accurate 3D reconstructions from endoscopic video is a promising avenue for longitudinal radiation-free analysis of sinus anatomy and surgical outcomes. Several methods for monocular reconstruction have been proposed, yielding visually pleasant 3D anatomical structures by retrieving relative camera poses with structure-from-motion-type algorithms and fusion of monocular depth estimates. However, due to the complex properties of the underlying algorithms and endoscopic scenes, the reconstruction pipeline may perform poorly or fail unexpectedly. Further, acquiring medical data conveys additional challenges, presenting difficulties in quantitatively benchmarking these models, understanding failure cases, and identifying critical components that contribute to their precision. In this work, we perform a quantitative analysis of a self-supervised approach for sinus reconstruction using endoscopic sequences paired with optical tracking and high-resolution computed tomography acquired from nine ex-vivo specimens. Our results show that the generated reconstructions are in high agreement with the anatomy, yielding an average point-to-mesh error of 0.91 mm between reconstructions and CT segmentations. However, in a point-to-point matching scenario, relevant for endoscope tracking and navigation, we found average target registration errors of 6.58 mm. We identified that pose and depth estimation inaccuracies contribute equally to this error and that locally consistent sequences with shorter trajectories generate more accurate reconstructions. These results suggest that achieving global consistency between relative camera poses and estimated depths with the anatomy is essential. In doing so, we can ensure proper synergy between all components of the pipeline for improved reconstructions that will facilitate clinical application of this innovative technology.
Eye Tracking for Tele-robotic Surgery: A Comparative Evaluation of Head-worn Solutions
Oct 18, 2023Abstract:Purpose: Metrics derived from eye-gaze-tracking and pupillometry show promise for cognitive load assessment, potentially enhancing training and patient safety through user-specific feedback in tele-robotic surgery. However, current eye-tracking solutions' effectiveness in tele-robotic surgery is uncertain compared to everyday situations due to close-range interactions causing extreme pupil angles and occlusions. To assess the effectiveness of modern eye-gaze-tracking solutions in tele-robotic surgery, we compare the Tobii Pro 3 Glasses and Pupil Labs Core, evaluating their pupil diameter and gaze stability when integrated with the da Vinci Research Kit (dVRK). Methods: The study protocol includes a nine-point gaze calibration followed by pick-and-place task using the dVRK and is repeated three times. After a final calibration, users view a 3x3 grid of AprilTags, focusing on each marker for 10 seconds, to evaluate gaze stability across dVRK-screen positions with the L2-norm. Different gaze calibrations assess calibration's temporal deterioration due to head movements. Pupil diameter stability is evaluated using the FFT from the pupil diameter during the pick-and-place tasks. Users perform this routine with both head-worn eye-tracking systems. Results: Data collected from ten users indicate comparable pupil diameter stability. FFTs of pupil diameters show similar amplitudes in high-frequency components. Tobii Glasses show more temporal gaze stability compared to Pupil Labs, though both eye trackers yield a similar 4cm error in gaze estimation without an outdated calibration. Conclusion: Both eye trackers demonstrate similar stability of the pupil diameter and gaze, when the calibration is not outdated, indicating comparable eye-tracking and pupillometry performance in tele-robotic surgery settings.
An Uncertainty-Driven GCN Refinement Strategy for Organ Segmentation
Dec 06, 2020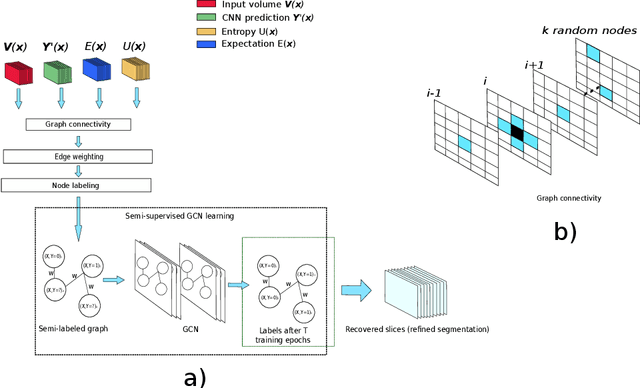
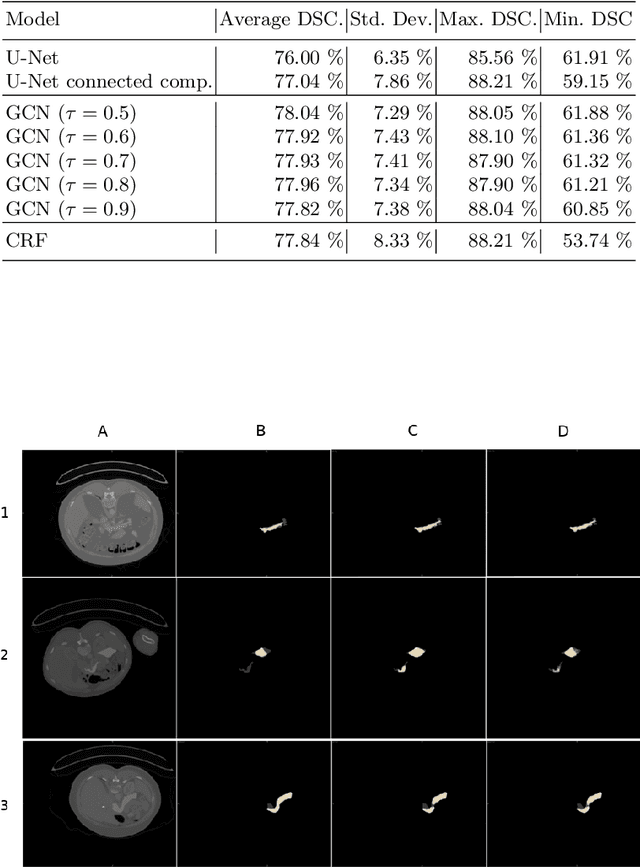
Abstract:Organ segmentation in CT volumes is an important pre-processing step in many computer assisted intervention and diagnosis methods. In recent years, convolutional neural networks have dominated the state of the art in this task. However, since this problem presents a challenging environment due to high variability in the organ's shape and similarity between tissues, the generation of false negative and false positive regions in the output segmentation is a common issue. Recent works have shown that the uncertainty analysis of the model can provide us with useful information about potential errors in the segmentation. In this context, we proposed a segmentation refinement method based on uncertainty analysis and graph convolutional networks. We employ the uncertainty levels of the convolutional network in a particular input volume to formulate a semi-supervised graph learning problem that is solved by training a graph convolutional network. To test our method we refine the initial output of a 2D U-Net. We validate our framework with the NIH pancreas dataset and the spleen dataset of the medical segmentation decathlon. We show that our method outperforms the state-of-the-art CRF refinement method by improving the dice score by 1% for the pancreas and 2% for spleen, with respect to the original U-Net's prediction. Finally, we perform a sensitivity analysis on the parameters of our proposal and discuss the applicability to other CNN architectures, the results, and current limitations of the model for future work in this research direction. For reproducibility purposes, we make our code publicly available at https://github.com/rodsom22/gcn_refinement.
Polyp-artifact relationship analysis using graph inductive learned representations
Sep 15, 2020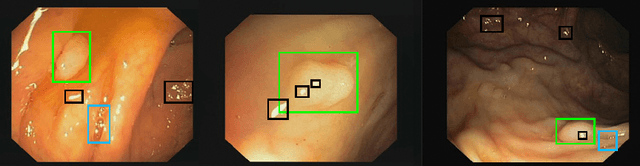
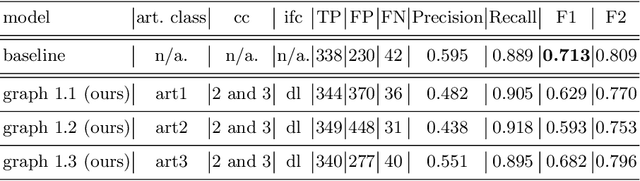
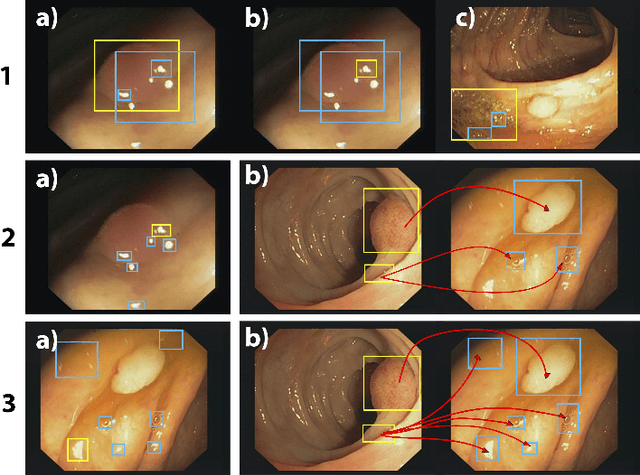
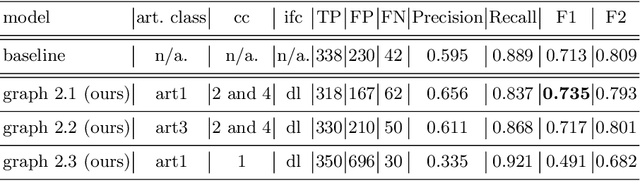
Abstract:The diagnosis process of colorectal cancer mainly focuses on the localization and characterization of abnormal growths in the colon tissue known as polyps. Despite recent advances in deep object localization, the localization of polyps remains challenging due to the similarities between tissues, and the high level of artifacts. Recent studies have shown the negative impact of the presence of artifacts in the polyp detection task, and have started to take them into account within the training process. However, the use of prior knowledge related to the spatial interaction of polyps and artifacts has not yet been considered. In this work, we incorporate artifact knowledge in a post-processing step. Our method models this task as an inductive graph representation learning problem, and is composed of training and inference steps. Detected bounding boxes around polyps and artifacts are considered as nodes connected by a defined criterion. The training step generates a node classifier with ground truth bounding boxes. In inference, we use this classifier to analyze a second graph, generated from artifact and polyp predictions given by region proposal networks. We evaluate how the choices in the connectivity and artifacts affect the performance of our method and show that it has the potential to reduce the false positives in the results of a region proposal network.
A learning without forgetting approach to incorporate artifact knowledge in polyp localization tasks
Feb 11, 2020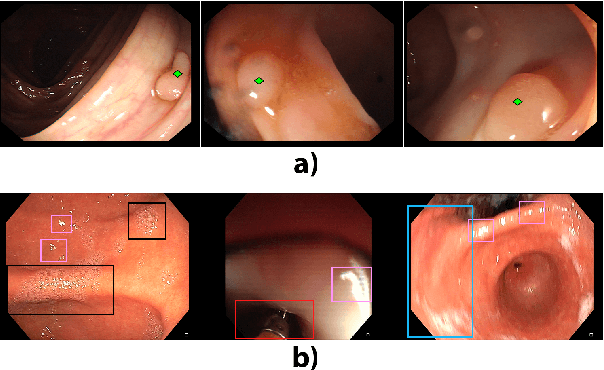
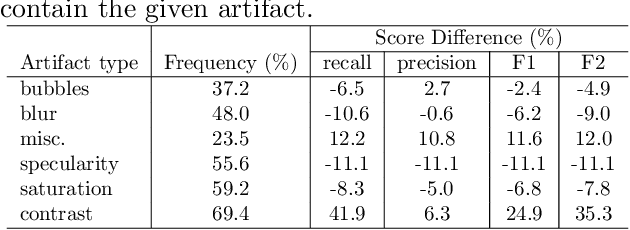
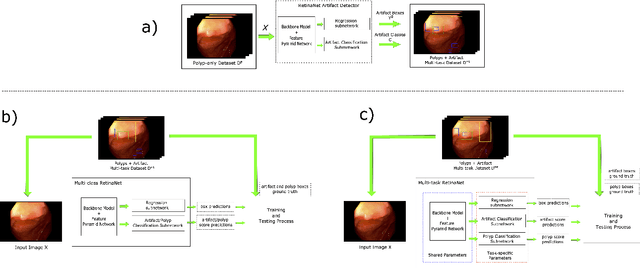
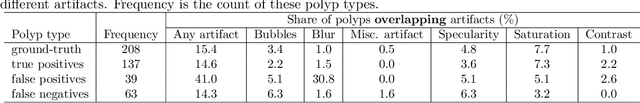
Abstract:Colorectal polyps are abnormalities in the colon tissue that can develop into colorectal cancer. The survival rate for patients is higher when the disease is detected at an early stage and polyps can be removed before they develop into malignant tumors. Deep learning methods have become the state of art in automatic polyp detection. However, the performance of current models heavily relies on the size and quality of the training datasets. Endoscopic video sequences tend to be corrupted by different artifacts affecting visibility and hence, the detection rates. In this work, we analyze the effects that artifacts have in the polyp localization problem. For this, we evaluate the RetinaNet architecture, originally defined for object localization. We also define a model inspired by the learning without forgetting framework, which allows us to employ artifact detection knowledge in the polyp localization problem. Finally, we perform several experiments to analyze the influence of the artifacts in the performance of these models. To our best knowledge, this is the first extensive analysis of the influence of artifact in polyp localization and the first work incorporating learning without forgetting ideas for simultaneous artifact and polyp localization tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge