Masaru Ishii
TwinOR: Photorealistic Digital Twins of Dynamic Operating Rooms for Embodied AI Research
Nov 10, 2025Abstract:Developing embodied AI for intelligent surgical systems requires safe, controllable environments for continual learning and evaluation. However, safety regulations and operational constraints in operating rooms (ORs) limit embodied agents from freely perceiving and interacting in realistic settings. Digital twins provide high-fidelity, risk-free environments for exploration and training. How we may create photorealistic and dynamic digital representations of ORs that capture relevant spatial, visual, and behavioral complexity remains unclear. We introduce TwinOR, a framework for constructing photorealistic, dynamic digital twins of ORs for embodied AI research. The system reconstructs static geometry from pre-scan videos and continuously models human and equipment motion through multi-view perception of OR activities. The static and dynamic components are fused into an immersive 3D environment that supports controllable simulation and embodied exploration. The proposed framework reconstructs complete OR geometry with centimeter level accuracy while preserving dynamic interaction across surgical workflows, enabling realistic renderings and a virtual playground for embodied AI systems. In our experiments, TwinOR simulates stereo and monocular sensor streams for geometry understanding and visual localization tasks. Models such as FoundationStereo and ORB-SLAM3 on TwinOR-synthesized data achieve performance within their reported accuracy on real indoor datasets, demonstrating that TwinOR provides sensor-level realism sufficient for perception and localization challenges. By establishing a real-to-sim pipeline for constructing dynamic, photorealistic digital twins of OR environments, TwinOR enables the safe, scalable, and data-efficient development and benchmarking of embodied AI, ultimately accelerating the deployment of embodied AI from sim-to-real.
Did you just see that? Arbitrary view synthesis for egocentric replay of operating room workflows from ambient sensors
Oct 06, 2025Abstract:Observing surgical practice has historically relied on fixed vantage points or recollections, leaving the egocentric visual perspectives that guide clinical decisions undocumented. Fixed-camera video can capture surgical workflows at the room-scale, but cannot reconstruct what each team member actually saw. Thus, these videos only provide limited insights into how decisions that affect surgical safety, training, and workflow optimization are made. Here we introduce EgoSurg, the first framework to reconstruct the dynamic, egocentric replays for any operating room (OR) staff directly from wall-mounted fixed-camera video, and thus, without intervention to clinical workflow. EgoSurg couples geometry-driven neural rendering with diffusion-based view enhancement, enabling high-visual fidelity synthesis of arbitrary and egocentric viewpoints at any moment. In evaluation across multi-site surgical cases and controlled studies, EgoSurg reconstructs person-specific visual fields and arbitrary viewpoints with high visual quality and fidelity. By transforming existing OR camera infrastructure into a navigable dynamic 3D record, EgoSurg establishes a new foundation for immersive surgical data science, enabling surgical practice to be visualized, experienced, and analyzed from every angle.
An Endoscopic Chisel: Intraoperative Imaging Carves 3D Anatomical Models
Feb 19, 2024Abstract:Purpose: Preoperative imaging plays a pivotal role in sinus surgery where CTs offer patient-specific insights of complex anatomy, enabling real-time intraoperative navigation to complement endoscopy imaging. However, surgery elicits anatomical changes not represented in the preoperative model, generating an inaccurate basis for navigation during surgery progression. Methods: We propose a first vision-based approach to update the preoperative 3D anatomical model leveraging intraoperative endoscopic video for navigated sinus surgery where relative camera poses are known. We rely on comparisons of intraoperative monocular depth estimates and preoperative depth renders to identify modified regions. The new depths are integrated in these regions through volumetric fusion in a truncated signed distance function representation to generate an intraoperative 3D model that reflects tissue manipulation. Results: We quantitatively evaluate our approach by sequentially updating models for a five-step surgical progression in an ex vivo specimen. We compute the error between correspondences from the updated model and ground-truth intraoperative CT in the region of anatomical modification. The resulting models show a decrease in error during surgical progression as opposed to increasing when no update is employed. Conclusion: Our findings suggest that preoperative 3D anatomical models can be updated using intraoperative endoscopy video in navigated sinus surgery. Future work will investigate improvements to monocular depth estimation as well as removing the need for external navigation systems. The resulting ability to continuously update the patient model may provide surgeons with a more precise understanding of the current anatomical state and paves the way toward a digital twin paradigm for sinus surgery.
A Quantitative Evaluation of Dense 3D Reconstruction of Sinus Anatomy from Monocular Endoscopic Video
Oct 22, 2023



Abstract:Generating accurate 3D reconstructions from endoscopic video is a promising avenue for longitudinal radiation-free analysis of sinus anatomy and surgical outcomes. Several methods for monocular reconstruction have been proposed, yielding visually pleasant 3D anatomical structures by retrieving relative camera poses with structure-from-motion-type algorithms and fusion of monocular depth estimates. However, due to the complex properties of the underlying algorithms and endoscopic scenes, the reconstruction pipeline may perform poorly or fail unexpectedly. Further, acquiring medical data conveys additional challenges, presenting difficulties in quantitatively benchmarking these models, understanding failure cases, and identifying critical components that contribute to their precision. In this work, we perform a quantitative analysis of a self-supervised approach for sinus reconstruction using endoscopic sequences paired with optical tracking and high-resolution computed tomography acquired from nine ex-vivo specimens. Our results show that the generated reconstructions are in high agreement with the anatomy, yielding an average point-to-mesh error of 0.91 mm between reconstructions and CT segmentations. However, in a point-to-point matching scenario, relevant for endoscope tracking and navigation, we found average target registration errors of 6.58 mm. We identified that pose and depth estimation inaccuracies contribute equally to this error and that locally consistent sequences with shorter trajectories generate more accurate reconstructions. These results suggest that achieving global consistency between relative camera poses and estimated depths with the anatomy is essential. In doing so, we can ensure proper synergy between all components of the pipeline for improved reconstructions that will facilitate clinical application of this innovative technology.
The Quiet Eye Phenomenon in Minimally Invasive Surgery
Sep 06, 2023Abstract:In this paper, we report our discovery of a gaze behavior called Quiet Eye (QE) in minimally invasive surgery. The QE behavior has been extensively studied in sports training and has been associated with higher level of expertise in multiple sports. We investigated the QE behavior in two independently collected data sets of surgeons performing tasks in a sinus surgery setting and a robotic surgery setting, respectively. Our results show that the QE behavior is more likely to occur in successful task executions and in performances of surgeons of high level of expertise. These results open the door to use the QE behavior in both training and skill assessment in minimally invasive surgery.
SAGE: SLAM with Appearance and Geometry Prior for Endoscopy
Feb 22, 2022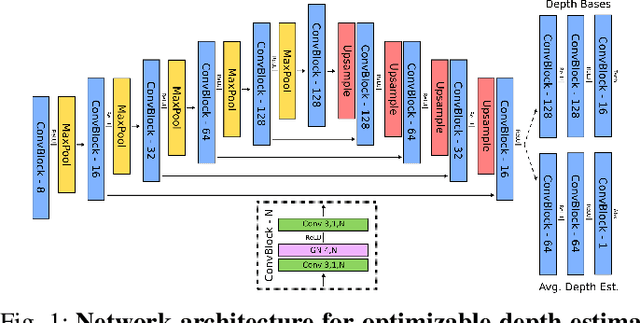
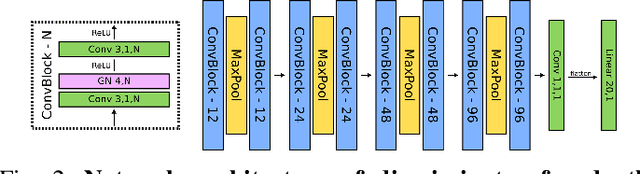
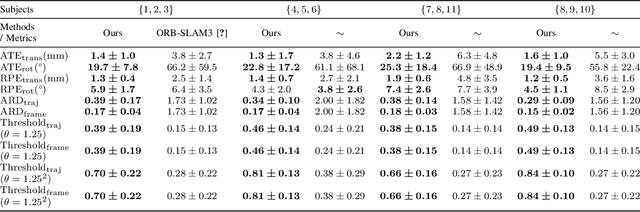
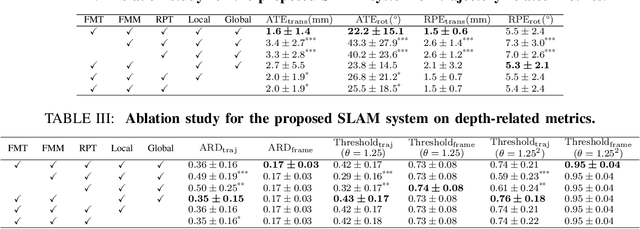
Abstract:In endoscopy, many applications (e.g., surgical navigation) would benefit from a real-time method that can simultaneously track the endoscope and reconstruct the dense 3D geometry of the observed anatomy from a monocular endoscopic video. To this end, we develop a Simultaneous Localization and Mapping system by combining the learning-based appearance and optimizable geometry priors and factor graph optimization. The appearance and geometry priors are explicitly learned in an end-to-end differentiable training pipeline to master the task of pair-wise image alignment, one of the core components of the SLAM system. In our experiments, the proposed SLAM system is shown to robustly handle the challenges of texture scarceness and illumination variation that are commonly seen in endoscopy. The system generalizes well to unseen endoscopes and subjects and performs favorably compared with a state-of-the-art feature-based SLAM system. The code repository is available at https://github.com/lppllppl920/SAGE-SLAM.git.
Learning Representations of Endoscopic Videos to Detect Tool Presence Without Supervision
Aug 27, 2020


Abstract:In this work, we explore whether it is possible to learn representations of endoscopic video frames to perform tasks such as identifying surgical tool presence without supervision. We use a maximum mean discrepancy (MMD) variational autoencoder (VAE) to learn low-dimensional latent representations of endoscopic videos and manipulate these representations to distinguish frames containing tools from those without tools. We use three different methods to manipulate these latent representations in order to predict tool presence in each frame. Our fully unsupervised methods can identify whether endoscopic video frames contain tools with average precision of 71.56, 73.93, and 76.18, respectively, comparable to supervised methods. Our code is available at https://github.com/zdavidli/tool-presence/
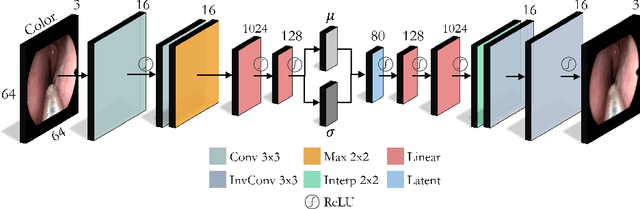
Extremely Dense Point Correspondences using a Learned Feature Descriptor
Mar 27, 2020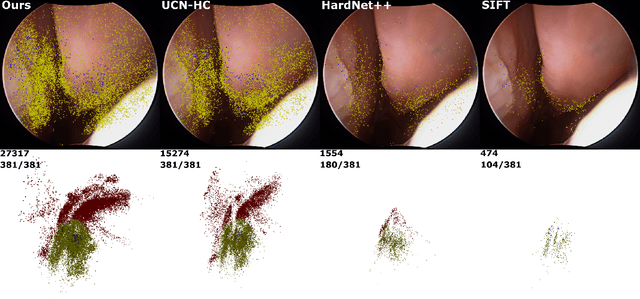
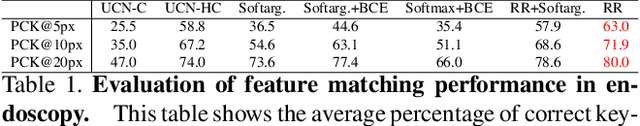
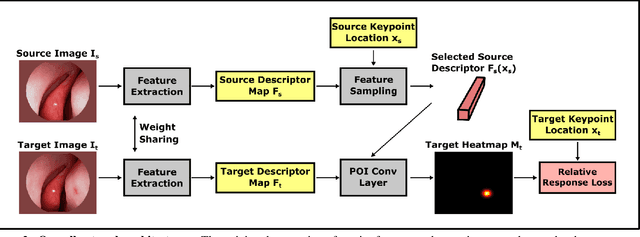
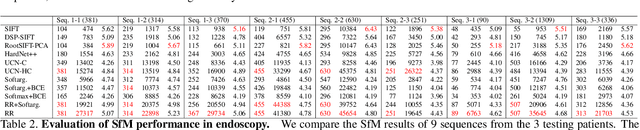
Abstract:High-quality 3D reconstructions from endoscopy video play an important role in many clinical applications, including surgical navigation where they enable direct video-CT registration. While many methods exist for general multi-view 3D reconstruction, these methods often fail to deliver satisfactory performance on endoscopic video. Part of the reason is that local descriptors that establish pair-wise point correspondences, and thus drive reconstruction, struggle when confronted with the texture-scarce surface of anatomy. Learning-based dense descriptors usually have larger receptive fields enabling the encoding of global information, which can be used to disambiguate matches. In this work, we present an effective self-supervised training scheme and novel loss design for dense descriptor learning. In direct comparison to recent local and dense descriptors on an in-house sinus endoscopy dataset, we demonstrate that our proposed dense descriptor can generalize to unseen patients and scopes, thereby largely improving the performance of Structure from Motion (SfM) in terms of model density and completeness. We also evaluate our method on a public dense optical flow dataset and a small-scale SfM public dataset to further demonstrate the effectiveness and generality of our method. The source code is available at https://github.com/lppllppl920/DenseDescriptorLearning-Pytorch.
Reconstructing Sinus Anatomy from Endoscopic Video -- Towards a Radiation-free Approach for Quantitative Longitudinal Assessment
Mar 18, 2020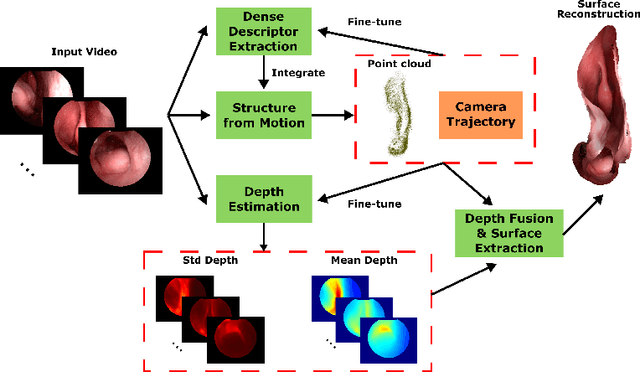
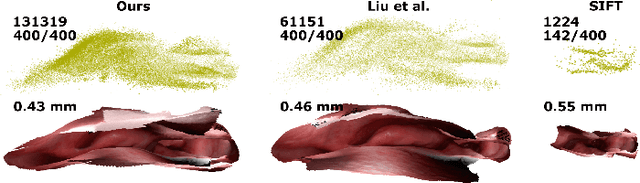
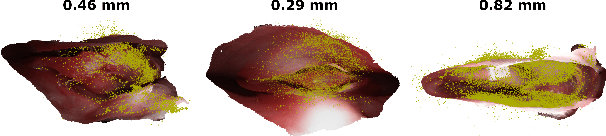
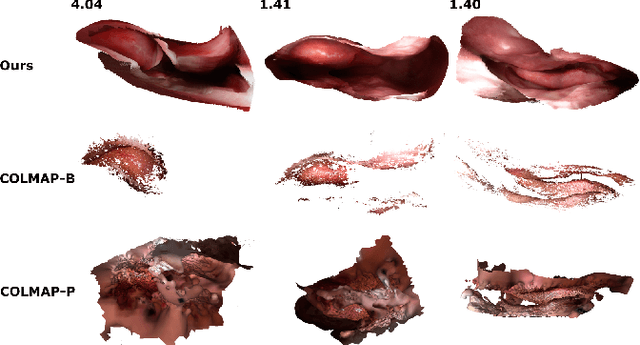
Abstract:Reconstructing accurate 3D surface models of sinus anatomy directly from an endoscopic video is a promising avenue for cross-sectional and longitudinal analysis to better understand the relationship between sinus anatomy and surgical outcomes. We present a patient-specific, learning-based method for 3D reconstruction of sinus surface anatomy directly and only from endoscopic videos. We demonstrate the effectiveness and accuracy of our method on in and ex vivo data where we compare to sparse reconstructions from Structure from Motion, dense reconstruction from COLMAP, and ground truth anatomy from CT. Our textured reconstructions are watertight and enable measurement of clinically relevant parameters in good agreement with CT. The source code will be made publicly available upon publication.
Self-supervised Dense 3D Reconstruction from Monocular Endoscopic Video
Sep 06, 2019
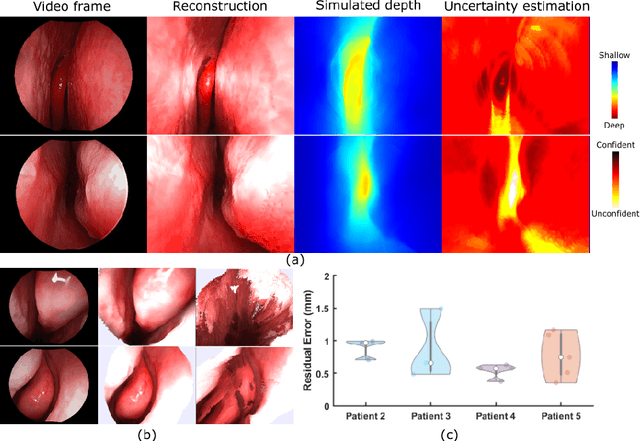
Abstract:We present a self-supervised learning-based pipeline for dense 3D reconstruction from full-length monocular endoscopic videos without a priori modeling of anatomy or shading. Our method only relies on unlabeled monocular endoscopic videos and conventional multi-view stereo algorithms, and requires neither manual interaction nor patient CT in both training and application phases. In a cross-patient study using CT scans as groundtruth, we show that our method is able to produce photo-realistic dense 3D reconstructions with submillimeter mean residual errors from endoscopic videos from unseen patients and scopes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge