Xingtong Liu
SAGE: SLAM with Appearance and Geometry Prior for Endoscopy
Feb 22, 2022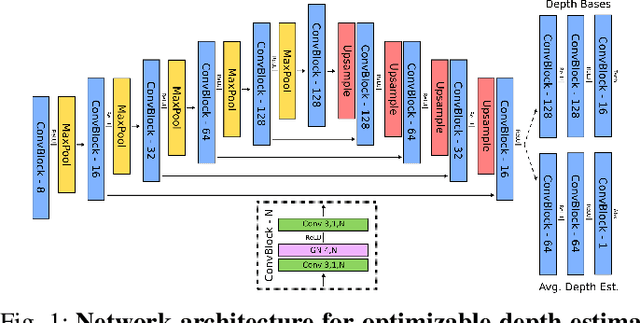
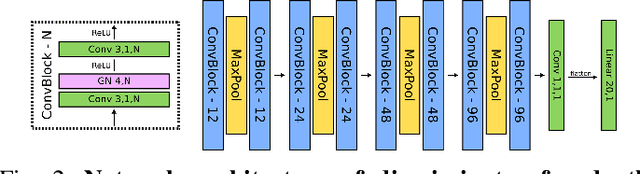
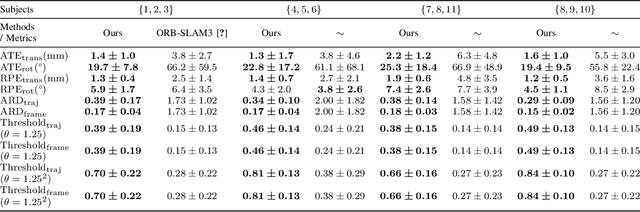
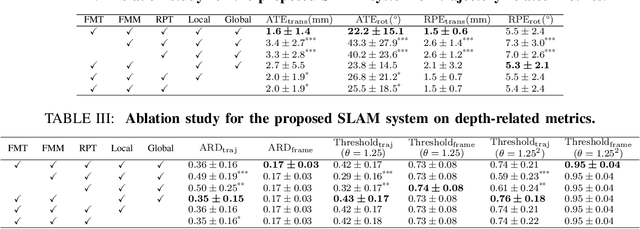
Abstract:In endoscopy, many applications (e.g., surgical navigation) would benefit from a real-time method that can simultaneously track the endoscope and reconstruct the dense 3D geometry of the observed anatomy from a monocular endoscopic video. To this end, we develop a Simultaneous Localization and Mapping system by combining the learning-based appearance and optimizable geometry priors and factor graph optimization. The appearance and geometry priors are explicitly learned in an end-to-end differentiable training pipeline to master the task of pair-wise image alignment, one of the core components of the SLAM system. In our experiments, the proposed SLAM system is shown to robustly handle the challenges of texture scarceness and illumination variation that are commonly seen in endoscopy. The system generalizes well to unseen endoscopes and subjects and performs favorably compared with a state-of-the-art feature-based SLAM system. The code repository is available at https://github.com/lppllppl920/SAGE-SLAM.git.
Revisiting Stereo Depth Estimation From a Sequence-to-Sequence Perspective with Transformers
Nov 06, 2020



Abstract:Stereo depth estimation relies on optimal correspondence matching between pixels on epipolar lines in the left and right image to infer depth. Rather than matching individual pixels, in this work, we revisit the problem from a sequence-to-sequence correspondence perspective to replace cost volume construction with dense pixel matching using position information and attention. This approach, named STereo TRansformer (STTR), has several advantages: It 1) relaxes the limitation of a fixed disparity range, 2) identifies occluded regions and provides confidence of estimation, and 3) imposes uniqueness constraints during the matching process. We report promising results on both synthetic and real-world datasets and demonstrate that STTR generalizes well across different domains, even without fine-tuning. Our code is publicly available at https://github.com/mli0603/stereo-transformer.
Extremely Dense Point Correspondences using a Learned Feature Descriptor
Mar 27, 2020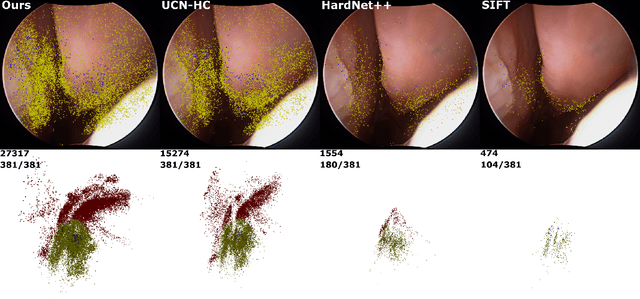
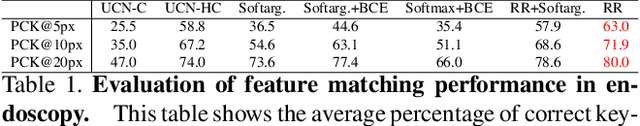
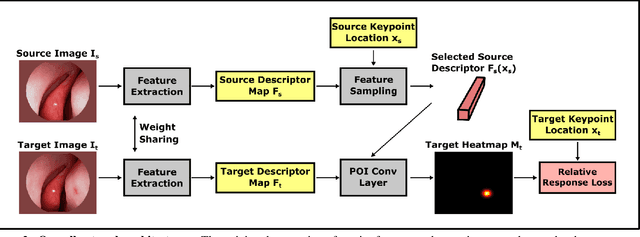
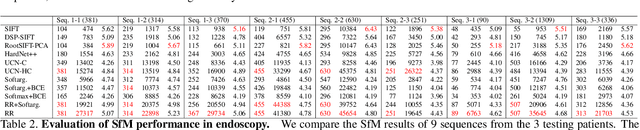
Abstract:High-quality 3D reconstructions from endoscopy video play an important role in many clinical applications, including surgical navigation where they enable direct video-CT registration. While many methods exist for general multi-view 3D reconstruction, these methods often fail to deliver satisfactory performance on endoscopic video. Part of the reason is that local descriptors that establish pair-wise point correspondences, and thus drive reconstruction, struggle when confronted with the texture-scarce surface of anatomy. Learning-based dense descriptors usually have larger receptive fields enabling the encoding of global information, which can be used to disambiguate matches. In this work, we present an effective self-supervised training scheme and novel loss design for dense descriptor learning. In direct comparison to recent local and dense descriptors on an in-house sinus endoscopy dataset, we demonstrate that our proposed dense descriptor can generalize to unseen patients and scopes, thereby largely improving the performance of Structure from Motion (SfM) in terms of model density and completeness. We also evaluate our method on a public dense optical flow dataset and a small-scale SfM public dataset to further demonstrate the effectiveness and generality of our method. The source code is available at https://github.com/lppllppl920/DenseDescriptorLearning-Pytorch.
Generalizing Spatial Transformers to Projective Geometry with Applications to 2D/3D Registration
Mar 24, 2020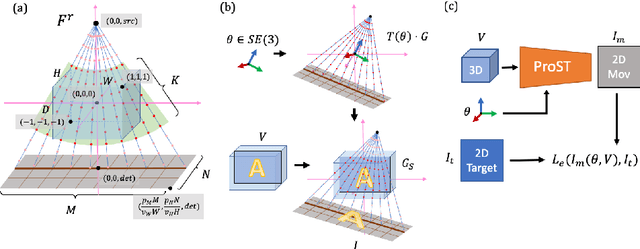
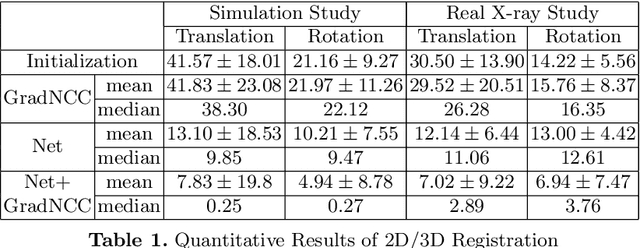
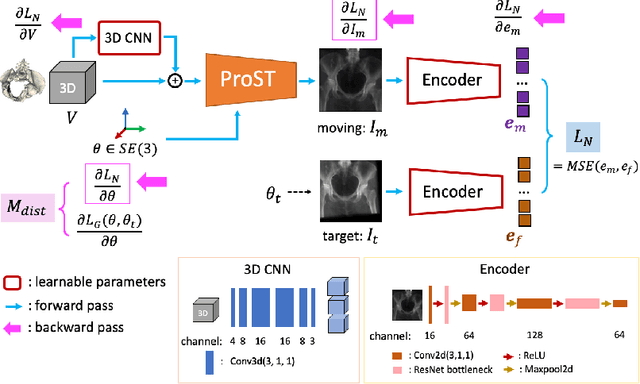
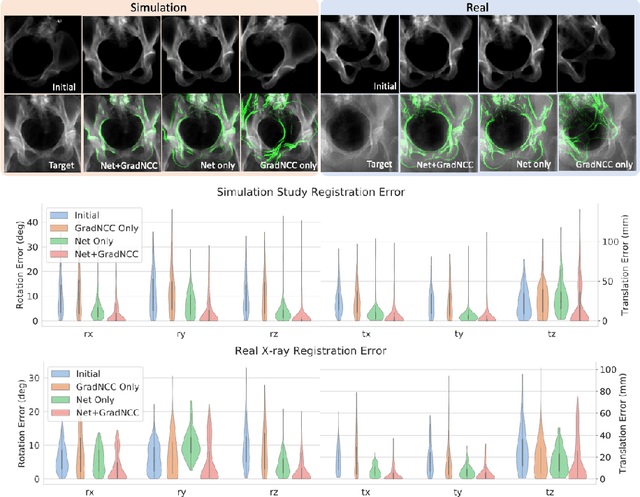
Abstract:Differentiable rendering is a technique to connect 3D scenes with corresponding 2D images. Since it is differentiable, processes during image formation can be learned. Previous approaches to differentiable rendering focus on mesh-based representations of 3D scenes, which is inappropriate for medical applications where volumetric, voxelized models are used to represent anatomy. We propose a novel Projective Spatial Transformer module that generalizes spatial transformers to projective geometry, thus enabling differentiable volume rendering. We demonstrate the usefulness of this architecture on the example of 2D/3D registration between radiographs and CT scans. Specifically, we show that our transformer enables end-to-end learning of an image processing and projection model that approximates an image similarity function that is convex with respect to the pose parameters, and can thus be optimized effectively using conventional gradient descent. To the best of our knowledge, this is the first time that spatial transformers have been described for projective geometry. The source code will be made public upon publication of this manuscript and we hope that our developments will benefit related 3D research applications.
Reconstructing Sinus Anatomy from Endoscopic Video -- Towards a Radiation-free Approach for Quantitative Longitudinal Assessment
Mar 18, 2020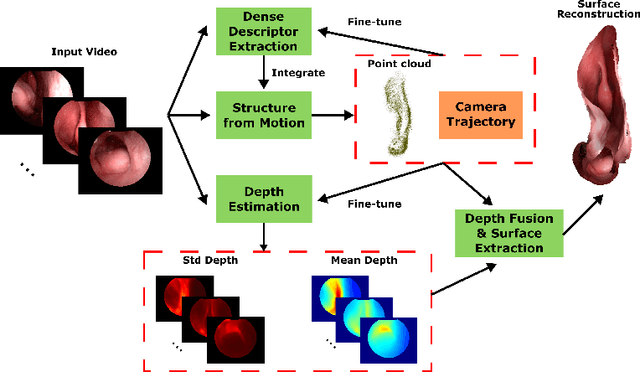
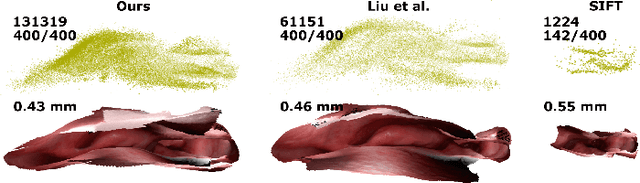
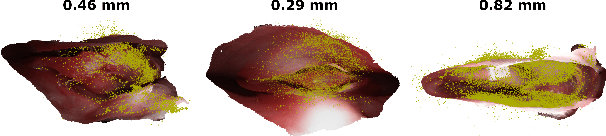
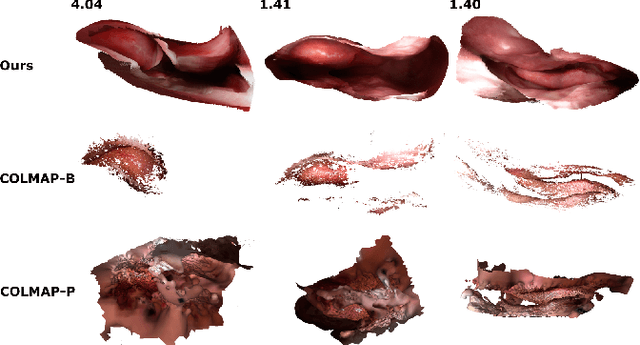
Abstract:Reconstructing accurate 3D surface models of sinus anatomy directly from an endoscopic video is a promising avenue for cross-sectional and longitudinal analysis to better understand the relationship between sinus anatomy and surgical outcomes. We present a patient-specific, learning-based method for 3D reconstruction of sinus surface anatomy directly and only from endoscopic videos. We demonstrate the effectiveness and accuracy of our method on in and ex vivo data where we compare to sparse reconstructions from Structure from Motion, dense reconstruction from COLMAP, and ground truth anatomy from CT. Our textured reconstructions are watertight and enable measurement of clinically relevant parameters in good agreement with CT. The source code will be made publicly available upon publication.
From Perspective X-ray Imaging to Parallax-Robust Orthographic Stitching
Mar 05, 2020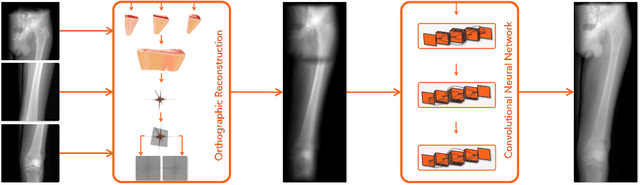
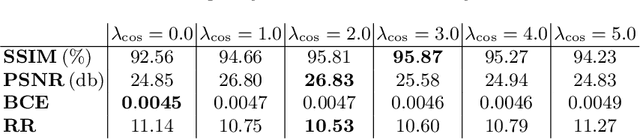
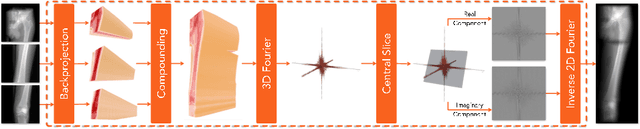
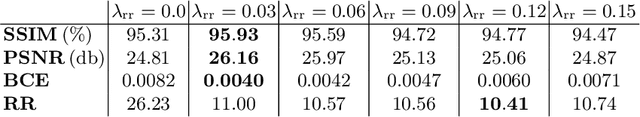
Abstract:Stitching images acquired under perspective projective geometry is a relevant topic in computer vision with multiple applications ranging from smartphone panoramas to the construction of digital maps. Image stitching is an equally prominent challenge in medical imaging, where the limited field-of-view captured by single images prohibits holistic analysis of patient anatomy. The barrier that prevents straight-forward mosaicing of 2D images is depth mismatch due to parallax. In this work, we leverage the Fourier slice theorem to aggregate information from multiple transmission images in parallax-free domains using fundamental principles of X-ray image formation. The semantics of the stitched image are restored using a novel deep learning strategy that exploits similarity measures designed around frequency, as well as dense and sparse spatial image content. Our pipeline, not only stitches images, but also provides orthographic reconstruction that enables metric measurements of clinically relevant quantities directly on the 2D image plane.
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020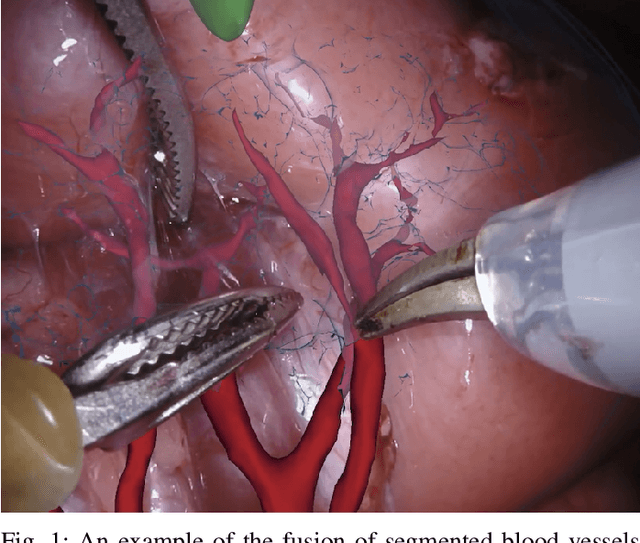
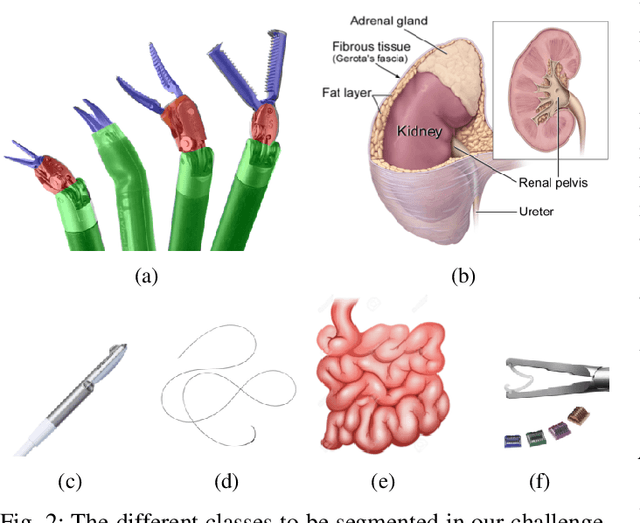
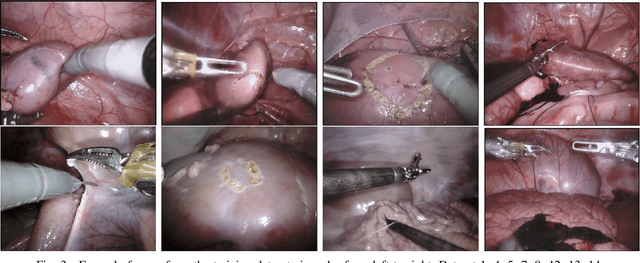
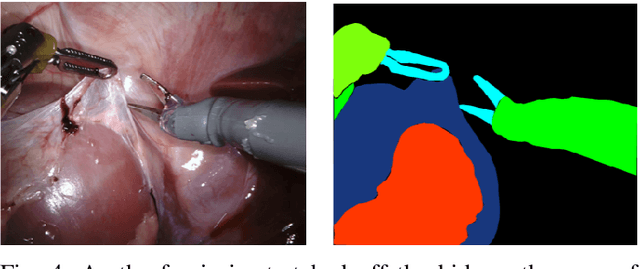
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
Self-supervised Dense 3D Reconstruction from Monocular Endoscopic Video
Sep 06, 2019
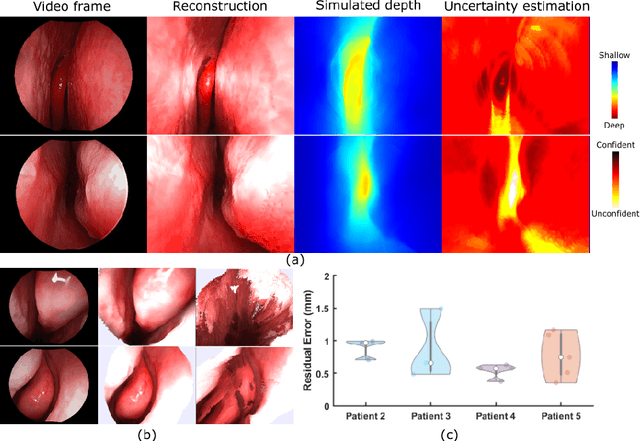
Abstract:We present a self-supervised learning-based pipeline for dense 3D reconstruction from full-length monocular endoscopic videos without a priori modeling of anatomy or shading. Our method only relies on unlabeled monocular endoscopic videos and conventional multi-view stereo algorithms, and requires neither manual interaction nor patient CT in both training and application phases. In a cross-patient study using CT scans as groundtruth, we show that our method is able to produce photo-realistic dense 3D reconstructions with submillimeter mean residual errors from endoscopic videos from unseen patients and scopes.
Self-supervised Learning for Dense Depth Estimation in Monocular Endoscopy
Feb 20, 2019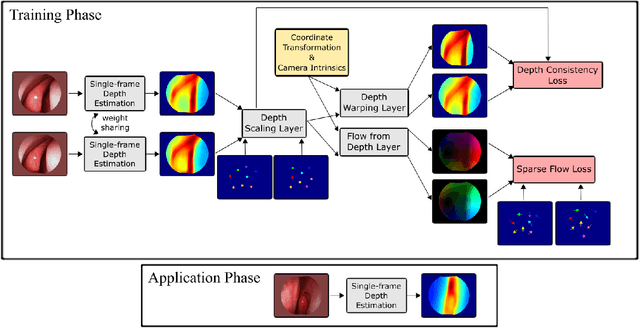
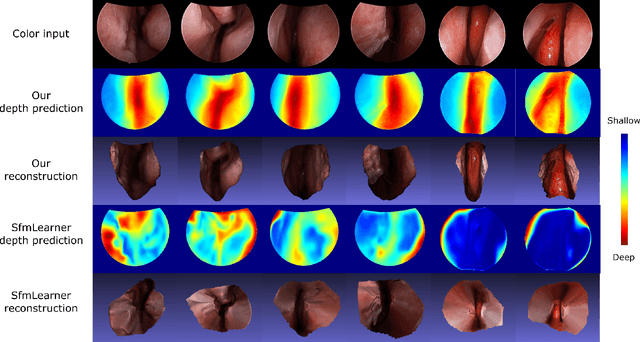
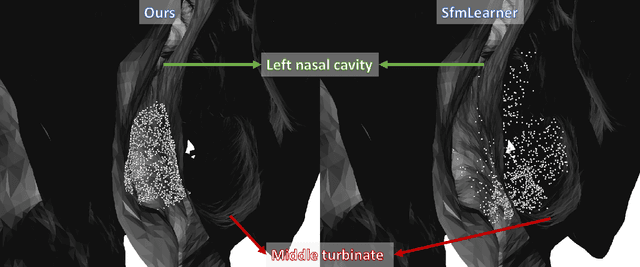
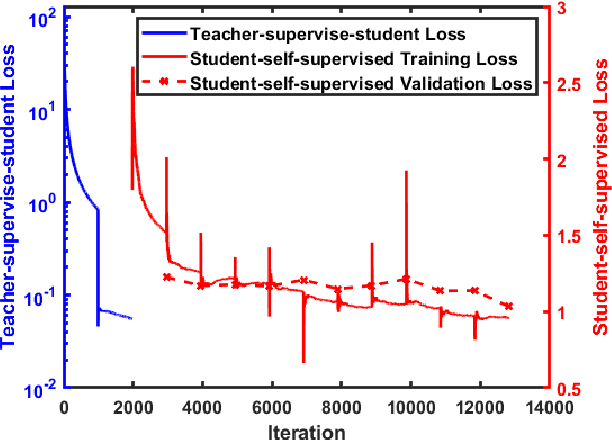
Abstract:We present a self-supervised approach to training convolutional neural networks for dense depth estimation from monocular endoscopy data without a priori modeling of anatomy or shading. Our method only requires monocular endoscopic video and a multi-view stereo method, e.g. structure from motion, to supervise learning in a sparse manner. Consequently, our method requires neither manual labeling nor patient computed tomography (CT) scan in the training and application phases. In a cross-patient experiment using CT scans as groundtruth, the proposed method achieved submillimeter root mean squared error. In a comparison study to a recent self-supervised depth estimation method designed for natural video on in vivo sinus endoscopy data, we demonstrate that the proposed approach outperforms the previous method by a large margin. The source code for this work is publicly available online at https://github.com/lppllppl920/EndoscopyDepthEstimation-Pytorch.
Learning to See Forces: Surgical Force Prediction with RGB-Point Cloud Temporal Convolutional Networks
Jul 31, 2018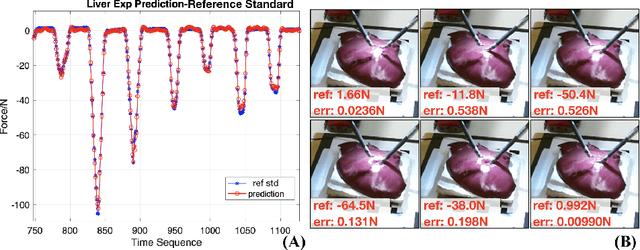
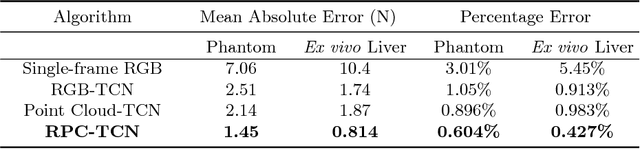
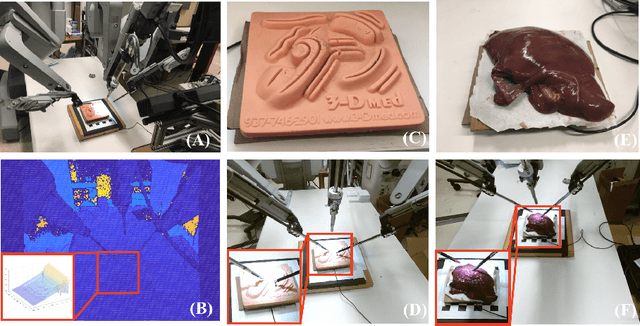
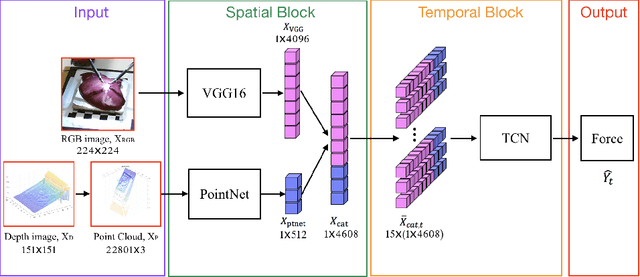
Abstract:Robotic surgery has been proven to offer clear advantages during surgical procedures, however, one of the major limitations is obtaining haptic feedback. Since it is often challenging to devise a hardware solution with accurate force feedback, we propose the use of "visual cues" to infer forces from tissue deformation. Endoscopic video is a passive sensor that is freely available, in the sense that any minimally-invasive procedure already utilizes it. To this end, we employ deep learning to infer forces from video as an attractive low-cost and accurate alternative to typically complex and expensive hardware solutions. First, we demonstrate our approach in a phantom setting using the da Vinci Surgical System affixed with an OptoForce sensor. Second, we then validate our method on an ex vivo liver organ. Our method results in a mean absolute error of 0.814 N in the ex vivo study, suggesting that it may be a promising alternative to hardware based surgical force feedback in endoscopic procedures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge