Ayushi Sinha
Department of Computer Science, The Johns Hopkins University, Baltimore, MD
Learning Representations of Endoscopic Videos to Detect Tool Presence Without Supervision
Aug 27, 2020
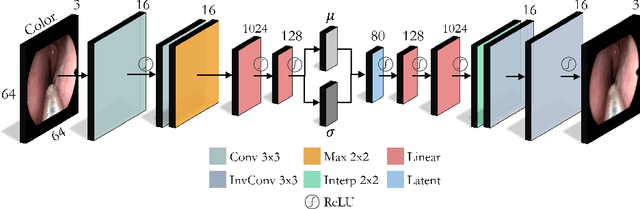


Abstract:In this work, we explore whether it is possible to learn representations of endoscopic video frames to perform tasks such as identifying surgical tool presence without supervision. We use a maximum mean discrepancy (MMD) variational autoencoder (VAE) to learn low-dimensional latent representations of endoscopic videos and manipulate these representations to distinguish frames containing tools from those without tools. We use three different methods to manipulate these latent representations in order to predict tool presence in each frame. Our fully unsupervised methods can identify whether endoscopic video frames contain tools with average precision of 71.56, 73.93, and 76.18, respectively, comparable to supervised methods. Our code is available at https://github.com/zdavidli/tool-presence/
Self-supervised Dense 3D Reconstruction from Monocular Endoscopic Video
Sep 06, 2019
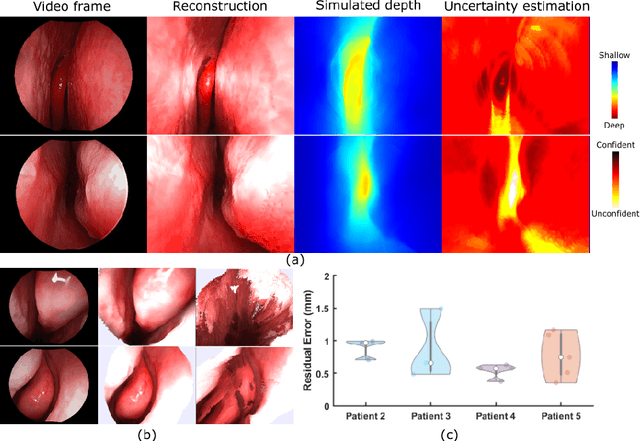
Abstract:We present a self-supervised learning-based pipeline for dense 3D reconstruction from full-length monocular endoscopic videos without a priori modeling of anatomy or shading. Our method only relies on unlabeled monocular endoscopic videos and conventional multi-view stereo algorithms, and requires neither manual interaction nor patient CT in both training and application phases. In a cross-patient study using CT scans as groundtruth, we show that our method is able to produce photo-realistic dense 3D reconstructions with submillimeter mean residual errors from endoscopic videos from unseen patients and scopes.
Self-supervised Learning for Dense Depth Estimation in Monocular Endoscopy
Feb 20, 2019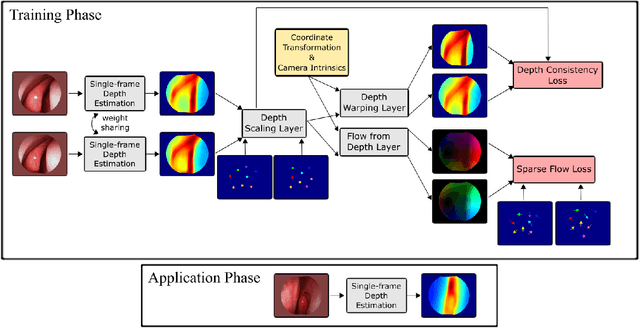
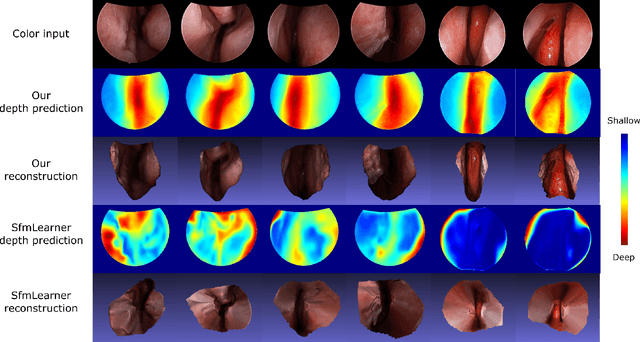
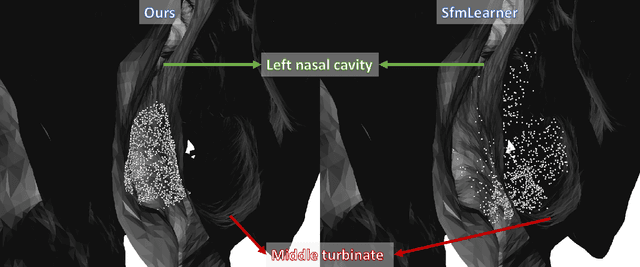
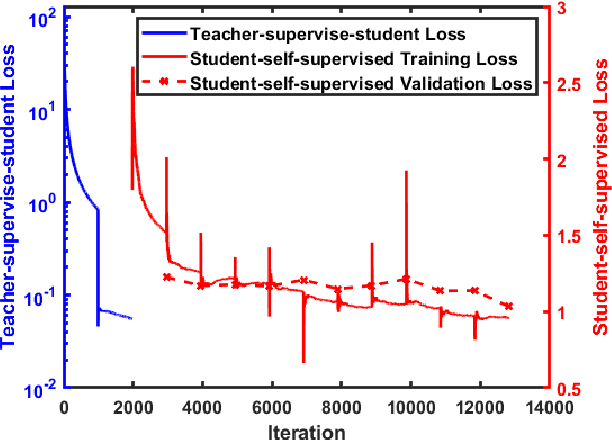
Abstract:We present a self-supervised approach to training convolutional neural networks for dense depth estimation from monocular endoscopy data without a priori modeling of anatomy or shading. Our method only requires monocular endoscopic video and a multi-view stereo method, e.g. structure from motion, to supervise learning in a sparse manner. Consequently, our method requires neither manual labeling nor patient computed tomography (CT) scan in the training and application phases. In a cross-patient experiment using CT scans as groundtruth, the proposed method achieved submillimeter root mean squared error. In a comparison study to a recent self-supervised depth estimation method designed for natural video on in vivo sinus endoscopy data, we demonstrate that the proposed approach outperforms the previous method by a large margin. The source code for this work is publicly available online at https://github.com/lppllppl920/EndoscopyDepthEstimation-Pytorch.
Towards automatic initialization of registration algorithms using simulated endoscopy images
Jun 28, 2018



Abstract:Registering images from different modalities is an active area of research in computer aided medical interventions. Several registration algorithms have been developed, many of which achieve high accuracy. However, these results are dependent on many factors, including the quality of the extracted features or segmentations being registered as well as the initial alignment. Although several methods have been developed towards improving segmentation algorithms and automating the segmentation process, few automatic initialization algorithms have been explored. In many cases, the initial alignment from which a registration is initiated is performed manually, which interferes with the clinical workflow. Our aim is to use scene classification in endoscopic procedures to achieve coarse alignment of the endoscope and a preoperative image of the anatomy. In this paper, we show using simulated scenes that a neural network can predict the region of anatomy (with respect to a preoperative image) that the endoscope is located in by observing a single endoscopic video frame. With limited training and without any hyperparameter tuning, our method achieves an accuracy of 76.53 (+/-1.19)%. There are several avenues for improvement, making this a promising direction of research. Code is available at https://github.com/AyushiSinha/AutoInitialization.
Endoscopic navigation in the absence of CT imaging
Jun 08, 2018


Abstract:Clinical examinations that involve endoscopic exploration of the nasal cavity and sinuses often do not have a reference image to provide structural context to the clinician. In this paper, we present a system for navigation during clinical endoscopic exploration in the absence of computed tomography (CT) scans by making use of shape statistics from past CT scans. Using a deformable registration algorithm along with dense reconstructions from video, we show that we are able to achieve submillimeter registrations in in-vivo clinical data and are able to assign confidence to these registrations using confidence criteria established using simulated data.
Anatomically Constrained Video-CT Registration via the V-IMLOP Algorithm
Oct 25, 2016



Abstract:Functional endoscopic sinus surgery (FESS) is a surgical procedure used to treat acute cases of sinusitis and other sinus diseases. FESS is fast becoming the preferred choice of treatment due to its minimally invasive nature. However, due to the limited field of view of the endoscope, surgeons rely on navigation systems to guide them within the nasal cavity. State of the art navigation systems report registration accuracy of over 1mm, which is large compared to the size of the nasal airways. We present an anatomically constrained video-CT registration algorithm that incorporates multiple video features. Our algorithm is robust in the presence of outliers. We also test our algorithm on simulated and in-vivo data, and test its accuracy against degrading initializations.
* 8 pages, 4 figures, MICCAI
Automatic Annotation of Axoplasmic Reticula in Pursuit of Connectomes using High-Resolution Neural EM Data
Apr 16, 2014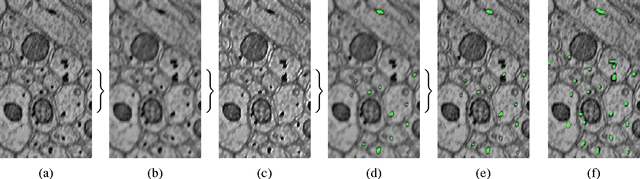

Abstract:Accurately estimating the wiring diagram of a brain, known as a connectome, at an ultrastructure level is an open research problem. Specifically, precisely tracking neural processes is difficult, especially across many image slices. Here, we propose a novel method to automatically identify and annotate small subcellular structures present in axons, known as axoplasmic reticula, through a 3D volume of high-resolution neural electron microscopy data. Our method produces high precision annotations, which can help improve automatic segmentation by using our results as seeds for segmentation, and as cues to aid segment merging.
Automatic Annotation of Axoplasmic Reticula in Pursuit of Connectomes
Apr 16, 2014

Abstract:In this paper, we present a new pipeline which automatically identifies and annotates axoplasmic reticula, which are small subcellular structures present only in axons. We run our algorithm on the Kasthuri11 dataset, which was color corrected using gradient-domain techniques to adjust contrast. We use a bilateral filter to smooth out the noise in this data while preserving edges, which highlights axoplasmic reticula. These axoplasmic reticula are then annotated using a morphological region growing algorithm. Additionally, we perform Laplacian sharpening on the bilaterally filtered data to enhance edges, and repeat the morphological region growing algorithm to annotate more axoplasmic reticula. We track our annotations through the slices to improve precision, and to create long objects to aid in segment merging. This method annotates axoplasmic reticula with high precision. Our algorithm can easily be adapted to annotate axoplasmic reticula in different sets of brain data by changing a few thresholds. The contribution of this work is the introduction of a straightforward and robust pipeline which annotates axoplasmic reticula with high precision, contributing towards advancements in automatic feature annotations in neural EM data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge