R. Jacob Vogelstein
Fast Neuromimetic Object Recognition using FPGA Outperforms GPU Implementations
Oct 31, 2015



Abstract:Recognition of objects in still images has traditionally been regarded as a difficult computational problem. Although modern automated methods for visual object recognition have achieved steadily increasing recognition accuracy, even the most advanced computational vision approaches are unable to obtain performance equal to that of humans. This has led to the creation of many biologically-inspired models of visual object recognition, among them the HMAX model. HMAX is traditionally known to achieve high accuracy in visual object recognition tasks at the expense of significant computational complexity. Increasing complexity, in turn, increases computation time, reducing the number of images that can be processed per unit time. In this paper we describe how the computationally intensive, biologically inspired HMAX model for visual object recognition can be modified for implementation on a commercial Field Programmable Gate Array, specifically the Xilinx Virtex 6 ML605 evaluation board with XC6VLX240T FPGA. We show that with minor modifications to the traditional HMAX model we can perform recognition on images of size 128x128 pixels at a rate of 190 images per second with a less than 1% loss in recognition accuracy in both binary and multi-class visual object recognition tasks.
* 14 pages, 8 figures, 5 tables
VESICLE: Volumetric Evaluation of Synaptic Interfaces using Computer vision at Large Scale
Sep 07, 2015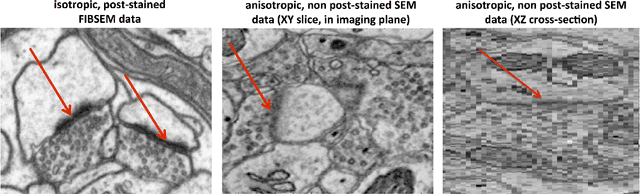
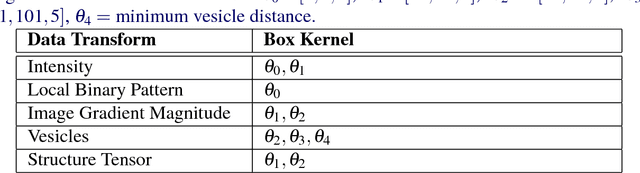
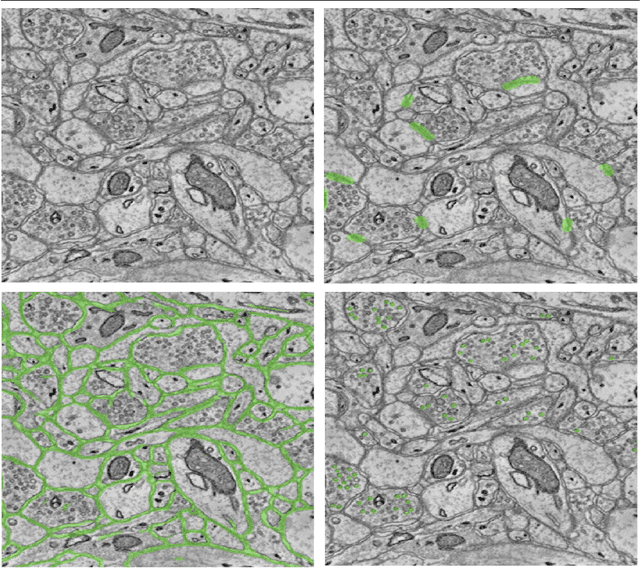
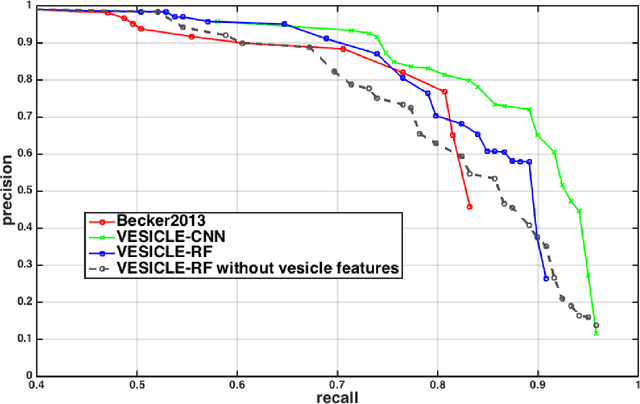
Abstract:An open challenge problem at the forefront of modern neuroscience is to obtain a comprehensive mapping of the neural pathways that underlie human brain function; an enhanced understanding of the wiring diagram of the brain promises to lead to new breakthroughs in diagnosing and treating neurological disorders. Inferring brain structure from image data, such as that obtained via electron microscopy (EM), entails solving the problem of identifying biological structures in large data volumes. Synapses, which are a key communication structure in the brain, are particularly difficult to detect due to their small size and limited contrast. Prior work in automated synapse detection has relied upon time-intensive biological preparations (post-staining, isotropic slice thicknesses) in order to simplify the problem. This paper presents VESICLE, the first known approach designed for mammalian synapse detection in anisotropic, non-post-stained data. Our methods explicitly leverage biological context, and the results exceed existing synapse detection methods in terms of accuracy and scalability. We provide two different approaches - one a deep learning classifier (VESICLE-CNN) and one a lightweight Random Forest approach (VESICLE-RF) to offer alternatives in the performance-scalability space. Addressing this synapse detection challenge enables the analysis of high-throughput imaging data soon expected to reach petabytes of data, and provide tools for more rapid estimation of brain-graphs. Finally, to facilitate community efforts, we developed tools for large-scale object detection, and demonstrated this framework to find $\approx$ 50,000 synapses in 60,000 $\mu m ^3$ (220 GB on disk) of electron microscopy data.
* v4: added clarifying figures and updates for readability. v3: fixed metadata. 11 pp v2: Added CNN classifier, significant changes to improve performance and generalization
An Automated Images-to-Graphs Framework for High Resolution Connectomics
Apr 30, 2015


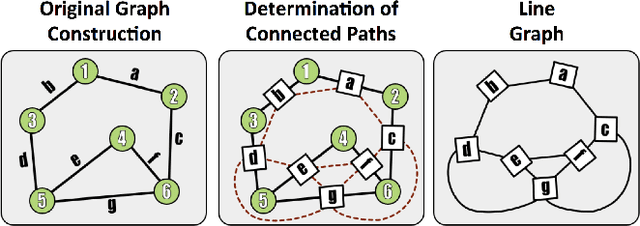
Abstract:Reconstructing a map of neuronal connectivity is a critical challenge in contemporary neuroscience. Recent advances in high-throughput serial section electron microscopy (EM) have produced massive 3D image volumes of nanoscale brain tissue for the first time. The resolution of EM allows for individual neurons and their synaptic connections to be directly observed. Recovering neuronal networks by manually tracing each neuronal process at this scale is unmanageable, and therefore researchers are developing automated image processing modules. Thus far, state-of-the-art algorithms focus only on the solution to a particular task (e.g., neuron segmentation or synapse identification). In this manuscript we present the first fully automated images-to-graphs pipeline (i.e., a pipeline that begins with an imaged volume of neural tissue and produces a brain graph without any human interaction). To evaluate overall performance and select the best parameters and methods, we also develop a metric to assess the quality of the output graphs. We evaluate a set of algorithms and parameters, searching possible operating points to identify the best available brain graph for our assessment metric. Finally, we deploy a reference end-to-end version of the pipeline on a large, publicly available data set. This provides a baseline result and framework for community analysis and future algorithm development and testing. All code and data derivatives have been made publicly available toward eventually unlocking new biofidelic computational primitives and understanding of neuropathologies.
Automatic Annotation of Axoplasmic Reticula in Pursuit of Connectomes
Apr 16, 2014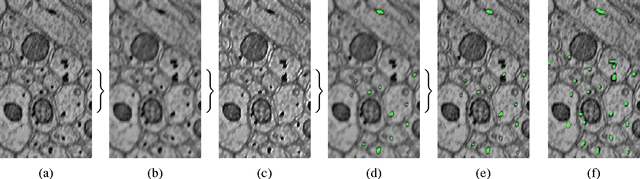

Abstract:In this paper, we present a new pipeline which automatically identifies and annotates axoplasmic reticula, which are small subcellular structures present only in axons. We run our algorithm on the Kasthuri11 dataset, which was color corrected using gradient-domain techniques to adjust contrast. We use a bilateral filter to smooth out the noise in this data while preserving edges, which highlights axoplasmic reticula. These axoplasmic reticula are then annotated using a morphological region growing algorithm. Additionally, we perform Laplacian sharpening on the bilaterally filtered data to enhance edges, and repeat the morphological region growing algorithm to annotate more axoplasmic reticula. We track our annotations through the slices to improve precision, and to create long objects to aid in segment merging. This method annotates axoplasmic reticula with high precision. Our algorithm can easily be adapted to annotate axoplasmic reticula in different sets of brain data by changing a few thresholds. The contribution of this work is the introduction of a straightforward and robust pipeline which annotates axoplasmic reticula with high precision, contributing towards advancements in automatic feature annotations in neural EM data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge