Ming Chuang
LivePose: Online 3D Reconstruction from Monocular Video with Dynamic Camera Poses
Mar 31, 2023



Abstract:Dense 3D reconstruction from RGB images traditionally assumes static camera pose estimates. This assumption has endured, even as recent works have increasingly focused on real-time methods for mobile devices. However, the assumption of one pose per image does not hold for online execution: poses from real-time SLAM are dynamic and may be updated following events such as bundle adjustment and loop closure. This has been addressed in the RGB-D setting, by de-integrating past views and re-integrating them with updated poses, but it remains largely untreated in the RGB-only setting. We formalize this problem to define the new task of online reconstruction from dynamically-posed images. To support further research, we introduce a dataset called LivePose containing the dynamic poses from a SLAM system running on ScanNet. We select three recent reconstruction systems and apply a framework based on de-integration to adapt each one to the dynamic-pose setting. In addition, we propose a novel, non-linear de-integration module that learns to remove stale scene content. We show that responding to pose updates is critical for high-quality reconstruction, and that our de-integration framework is an effective solution.
Automatic Annotation of Axoplasmic Reticula in Pursuit of Connectomes
Apr 16, 2014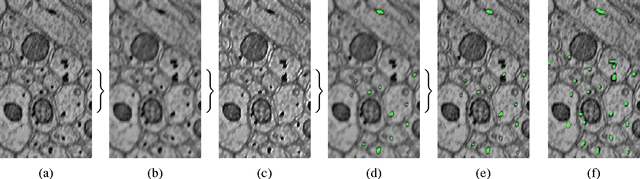

Abstract:In this paper, we present a new pipeline which automatically identifies and annotates axoplasmic reticula, which are small subcellular structures present only in axons. We run our algorithm on the Kasthuri11 dataset, which was color corrected using gradient-domain techniques to adjust contrast. We use a bilateral filter to smooth out the noise in this data while preserving edges, which highlights axoplasmic reticula. These axoplasmic reticula are then annotated using a morphological region growing algorithm. Additionally, we perform Laplacian sharpening on the bilaterally filtered data to enhance edges, and repeat the morphological region growing algorithm to annotate more axoplasmic reticula. We track our annotations through the slices to improve precision, and to create long objects to aid in segment merging. This method annotates axoplasmic reticula with high precision. Our algorithm can easily be adapted to annotate axoplasmic reticula in different sets of brain data by changing a few thresholds. The contribution of this work is the introduction of a straightforward and robust pipeline which annotates axoplasmic reticula with high precision, contributing towards advancements in automatic feature annotations in neural EM data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge