Michael Kazhdan
Department of Computer Science, The Johns Hopkins University, Baltimore, MD
A Novel Method to Improve Quality Surface Coverage in Multi-View Capture
Jul 21, 2024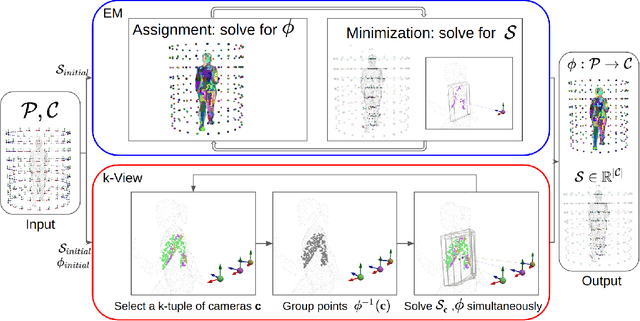
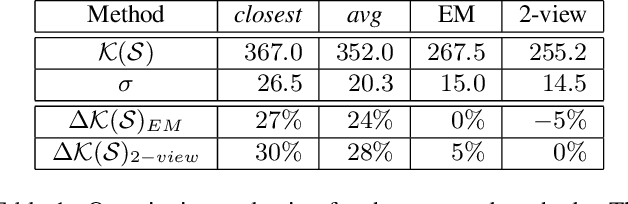
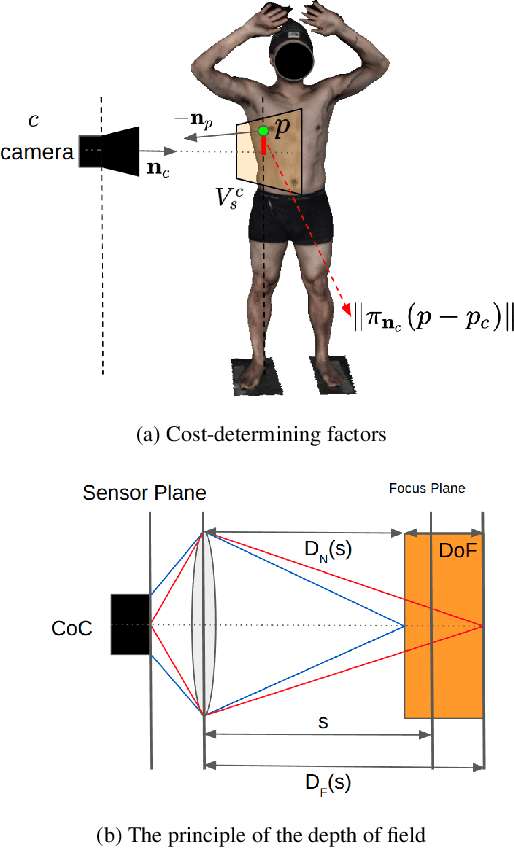
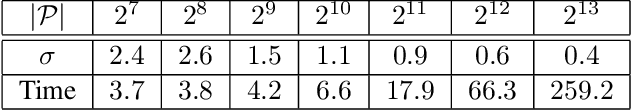
Abstract:The depth of field of a camera is a limiting factor for applications that require taking images at a short subject-to-camera distance or using a large focal length, such as total body photography, archaeology, and other close-range photogrammetry applications. Furthermore, in multi-view capture, where the target is larger than the camera's field of view, an efficient way to optimize surface coverage captured with quality remains a challenge. Given the 3D mesh of the target object and camera poses, we propose a novel method to derive a focus distance for each camera that optimizes the quality of the covered surface area. We first design an Expectation-Minimization (EM) algorithm to assign points on the mesh uniquely to cameras and then solve for a focus distance for each camera given the associated point set. We further improve the quality surface coverage by proposing a $k$-view algorithm that solves for the points assignment and focus distances by considering multiple views simultaneously. We demonstrate the effectiveness of the proposed method under various simulations for total body photography. The EM and $k$-view algorithms improve the relative cost of the baseline single-view methods by at least $24$% and $28$% respectively, corresponding to increasing the in-focus surface area by roughly $1550$ cm$^2$ and $1780$ cm$^2$. We believe the algorithms can be useful in a number of vision applications that require photogrammetric details but are limited by the depth of field.
Skin Lesion Correspondence Localization in Total Body Photography
Jul 18, 2023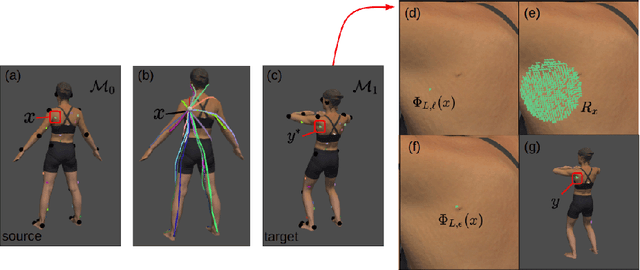

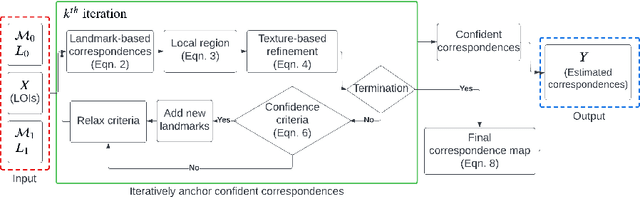
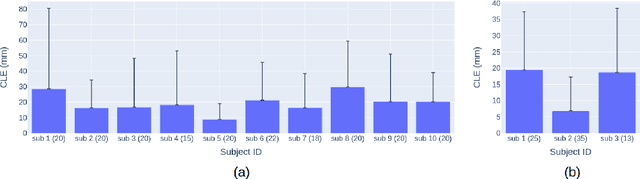
Abstract:Longitudinal tracking of skin lesions - finding correspondence, changes in morphology, and texture - is beneficial to the early detection of melanoma. However, it has not been well investigated in the context of full-body imaging. We propose a novel framework combining geometric and texture information to localize skin lesion correspondence from a source scan to a target scan in total body photography (TBP). Body landmarks or sparse correspondence are first created on the source and target 3D textured meshes. Every vertex on each of the meshes is then mapped to a feature vector characterizing the geodesic distances to the landmarks on that mesh. Then, for each lesion of interest (LOI) on the source, its corresponding location on the target is first coarsely estimated using the geometric information encoded in the feature vectors and then refined using the texture information. We evaluated the framework quantitatively on both a public and a private dataset, for which our success rates (at 10 mm criterion) are comparable to the only reported longitudinal study. As full-body 3D capture becomes more prevalent and has higher quality, we expect the proposed method to constitute a valuable step in the longitudinal tracking of skin lesions.
Improving Surgical Situational Awareness with Signed Distance Field: A Pilot Study in Virtual Reality
Mar 03, 2023Abstract:The introduction of image-guided surgical navigation (IGSN) has greatly benefited technically demanding surgical procedures by providing real-time support and guidance to the surgeon during surgery. To develop effective IGSN, a careful selection of the information provided to the surgeon is needed. However, identifying optimal feedback modalities is challenging due to the broad array of available options. To address this problem, we have developed an open-source library that facilitates the development of multimodal navigation systems in a wide range of surgical procedures relying on medical imaging data. To provide guidance, our system calculates the minimum distance between the surgical instrument and the anatomy and then presents this information to the user through different mechanisms. The real-time performance of our approach is achieved by calculating Signed Distance Fields at initialization from segmented anatomical volumes. Using this framework, we developed a multimodal surgical navigation system to help surgeons navigate anatomical variability in a skull-base surgery simulation environment. Three different feedback modalities were explored: visual, auditory, and haptic. To evaluate the proposed system, a pilot user study was conducted in which four clinicians performed mastoidectomy procedures with and without guidance. Each condition was assessed using objective performance and subjective workload metrics. This pilot user study showed improvements in procedural safety without additional time or workload. These results demonstrate our pipeline's successful use case in the context of mastoidectomy.
Möbius Convolutions for Spherical CNNs
Jan 28, 2022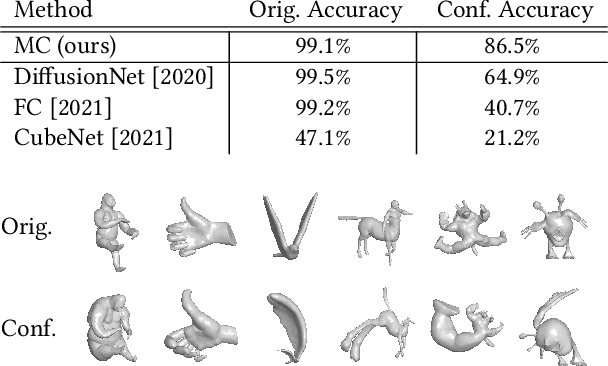
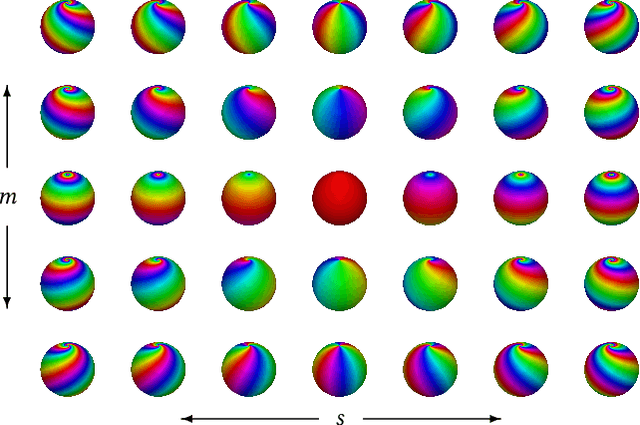

Abstract:M\"{o}bius transformations play an important role in both geometry and spherical image processing -- they are the group of conformal automorphisms of 2D surfaces and the spherical equivalent of homographies. Here we present a novel, M\"{o}bius-equivariant spherical convolution operator which we call M\"{o}bius convolution, and with it, develop the foundations for M\"{o}bius-equivariant spherical CNNs. Our approach is based on a simple observation: to achieve equivariance, we only need to consider the lower-dimensional subgroup which transforms the positions of points as seen in the frames of their neighbors. To efficiently compute M\"{o}bius convolutions at scale we derive an approximation of the action of the transformations on spherical filters, allowing us to compute our convolutions in the spectral domain with the fast Spherical Harmonic Transform. The resulting framework is both flexible and descriptive, and we demonstrate its utility by achieving promising results in both shape classification and image segmentation tasks.
Field Convolutions for Surface CNNs
Apr 08, 2021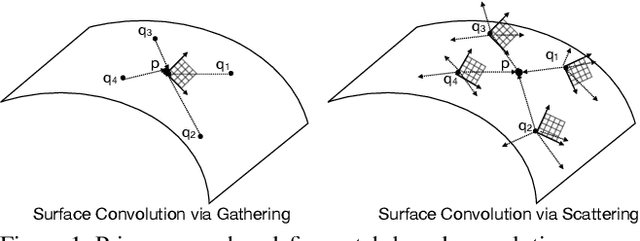
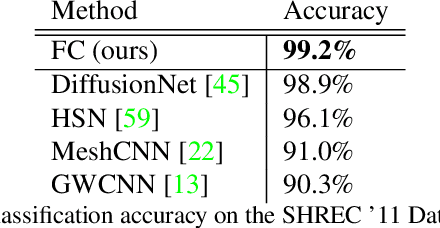
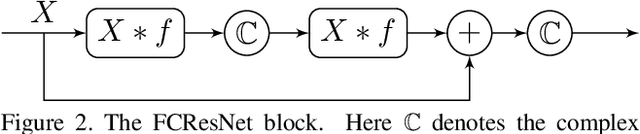
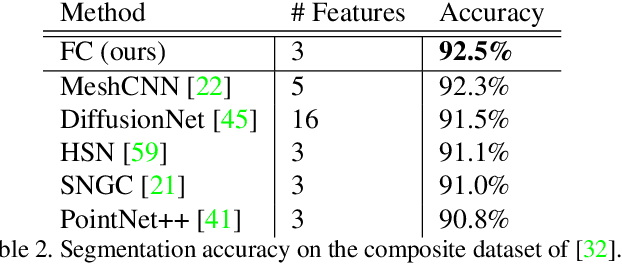
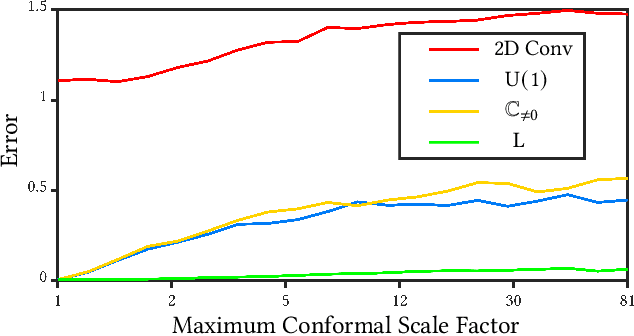
Abstract:We present a novel surface convolution operator acting on vector fields that is based on a simple observation: instead of combining neighboring features with respect to a single coordinate parameterization defined at a given point, we have every neighbor describe the position of the point within its own coordinate frame. This formulation combines intrinsic spatial convolution with parallel transport in a scattering operation while placing no constraints on the filters themselves, providing a definition of convolution that commutes with the action of isometries, has increased descriptive potential, and is robust to noise and other nuisance factors. The result is a rich notion of convolution which we call field convolution, well-suited for CNNs on surfaces. Field convolutions are flexible and straight-forward to implement, and their highly discriminating nature has cascading effects throughout the learning pipeline. Using simple networks constructed from residual field convolution blocks, we achieve state-of-the-art results on standard benchmarks in fundamental geometry processing tasks, such as shape classification, segmentation, correspondence, and sparse matching.
Efficient Spatially Adaptive Convolution and Correlation
Jun 23, 2020
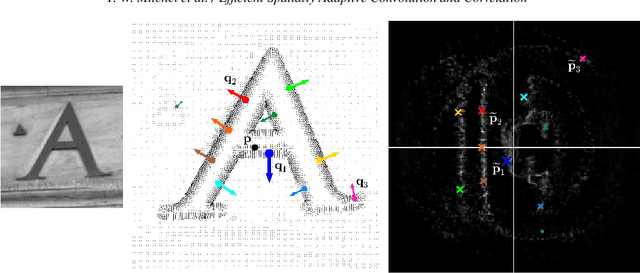
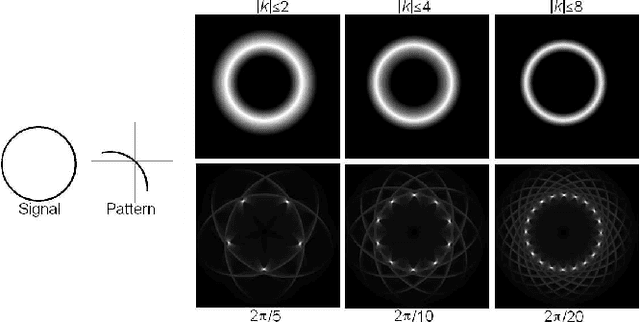
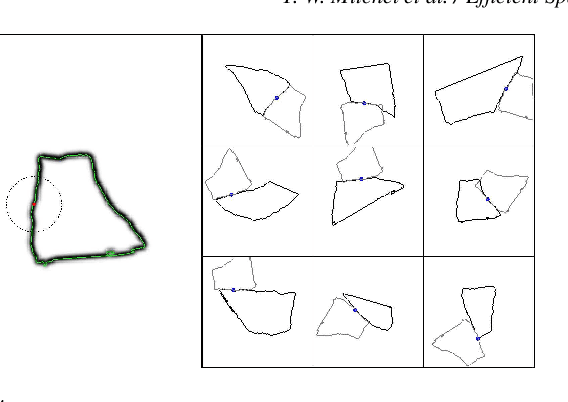
Abstract:Fast methods for convolution and correlation underlie a variety of applications in computer vision and graphics, including efficient filtering, analysis, and simulation. However, standard convolution and correlation are inherently limited to fixed filters: spatial adaptation is impossible without sacrificing efficient computation. In early work, Freeman and Adelson have shown how steerable filters can address this limitation, providing a way for rotating the filter as it is passed over the signal. In this work, we provide a general, representation-theoretic, framework that allows for spatially varying linear transformations to be applied to the filter. This framework allows for efficient implementation of extended convolution and correlation for transformation groups such as rotation (in 2D and 3D) and scale, and provides a new interpretation for previous methods including steerable filters and the generalized Hough transform. We present applications to pattern matching, image feature description, vector field visualization, and adaptive image filtering.
Automatic Annotation of Axoplasmic Reticula in Pursuit of Connectomes using High-Resolution Neural EM Data
Apr 16, 2014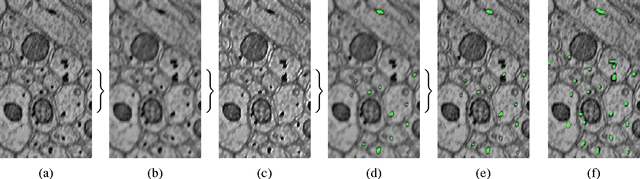

Abstract:Accurately estimating the wiring diagram of a brain, known as a connectome, at an ultrastructure level is an open research problem. Specifically, precisely tracking neural processes is difficult, especially across many image slices. Here, we propose a novel method to automatically identify and annotate small subcellular structures present in axons, known as axoplasmic reticula, through a 3D volume of high-resolution neural electron microscopy data. Our method produces high precision annotations, which can help improve automatic segmentation by using our results as seeds for segmentation, and as cues to aid segment merging.
Automatic Annotation of Axoplasmic Reticula in Pursuit of Connectomes
Apr 16, 2014

Abstract:In this paper, we present a new pipeline which automatically identifies and annotates axoplasmic reticula, which are small subcellular structures present only in axons. We run our algorithm on the Kasthuri11 dataset, which was color corrected using gradient-domain techniques to adjust contrast. We use a bilateral filter to smooth out the noise in this data while preserving edges, which highlights axoplasmic reticula. These axoplasmic reticula are then annotated using a morphological region growing algorithm. Additionally, we perform Laplacian sharpening on the bilaterally filtered data to enhance edges, and repeat the morphological region growing algorithm to annotate more axoplasmic reticula. We track our annotations through the slices to improve precision, and to create long objects to aid in segment merging. This method annotates axoplasmic reticula with high precision. Our algorithm can easily be adapted to annotate axoplasmic reticula in different sets of brain data by changing a few thresholds. The contribution of this work is the introduction of a straightforward and robust pipeline which annotates axoplasmic reticula with high precision, contributing towards advancements in automatic feature annotations in neural EM data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge