S. Swaroop Vedula
StepAL: Step-aware Active Learning for Cataract Surgical Videos
Jul 29, 2025Abstract:Active learning (AL) can reduce annotation costs in surgical video analysis while maintaining model performance. However, traditional AL methods, developed for images or short video clips, are suboptimal for surgical step recognition due to inter-step dependencies within long, untrimmed surgical videos. These methods typically select individual frames or clips for labeling, which is ineffective for surgical videos where annotators require the context of the entire video for annotation. To address this, we propose StepAL, an active learning framework designed for full video selection in surgical step recognition. StepAL integrates a step-aware feature representation, which leverages pseudo-labels to capture the distribution of predicted steps within each video, with an entropy-weighted clustering strategy. This combination prioritizes videos that are both uncertain and exhibit diverse step compositions for annotation. Experiments on two cataract surgery datasets (Cataract-1k and Cataract-101) demonstrate that StepAL consistently outperforms existing active learning approaches, achieving higher accuracy in step recognition with fewer labeled videos. StepAL offers an effective approach for efficient surgical video analysis, reducing the annotation burden in developing computer-assisted surgical systems.
$\mathsf{CSMAE~}$:~Cataract Surgical Masked Autoencoder (MAE) based Pre-training
Feb 12, 2025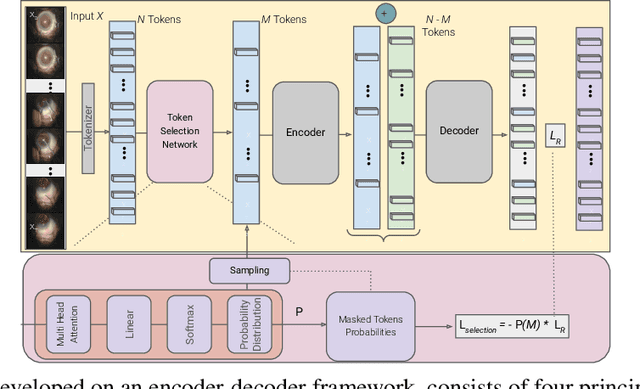
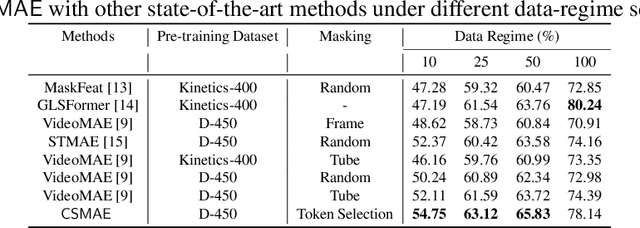

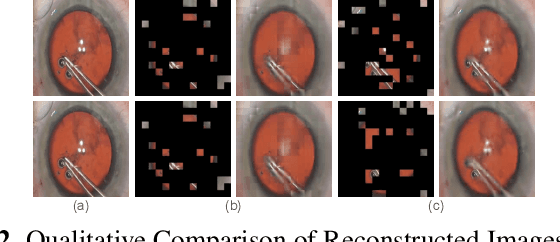
Abstract:Automated analysis of surgical videos is crucial for improving surgical training, workflow optimization, and postoperative assessment. We introduce a CSMAE, Masked Autoencoder (MAE)-based pretraining approach, specifically developed for Cataract Surgery video analysis, where instead of randomly selecting tokens for masking, they are selected based on the spatiotemporal importance of the token. We created a large dataset of cataract surgery videos to improve the model's learning efficiency and expand its robustness in low-data regimes. Our pre-trained model can be easily adapted for specific downstream tasks via fine-tuning, serving as a robust backbone for further analysis. Through rigorous testing on a downstream step-recognition task on two Cataract Surgery video datasets, D99 and Cataract-101, our approach surpasses current state-of-the-art self-supervised pretraining and adapter-based transfer learning methods by a significant margin. This advancement not only demonstrates the potential of our MAE-based pretraining in the field of surgical video analysis but also sets a new benchmark for future research.
Federated Black-Box Adaptation for Semantic Segmentation
Oct 31, 2024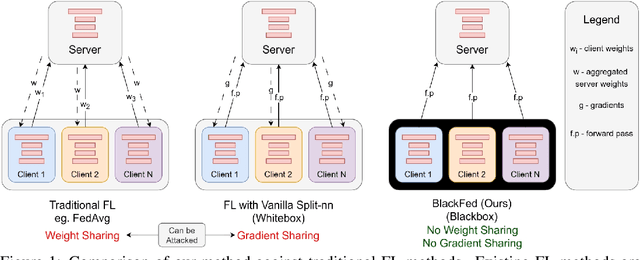
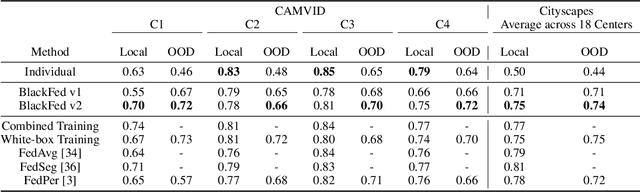
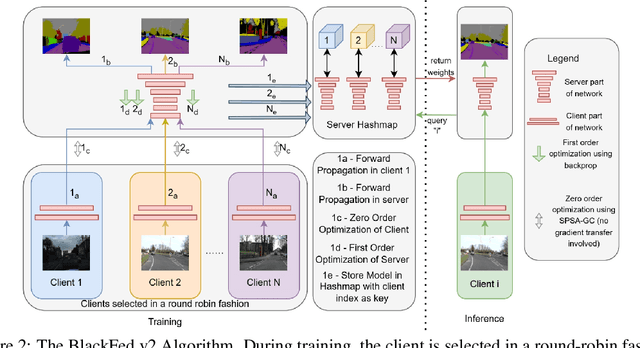
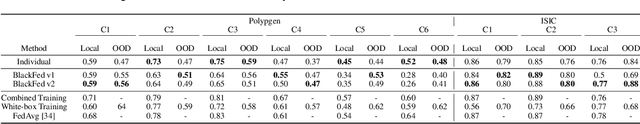
Abstract:Federated Learning (FL) is a form of distributed learning that allows multiple institutions or clients to collaboratively learn a global model to solve a task. This allows the model to utilize the information from every institute while preserving data privacy. However, recent studies show that the promise of protecting the privacy of data is not upheld by existing methods and that it is possible to recreate the training data from the different institutions. This is done by utilizing gradients transferred between the clients and the global server during training or by knowing the model architecture at the client end. In this paper, we propose a federated learning framework for semantic segmentation without knowing the model architecture nor transferring gradients between the client and the server, thus enabling better privacy preservation. We propose BlackFed - a black-box adaptation of neural networks that utilizes zero order optimization (ZOO) to update the client model weights and first order optimization (FOO) to update the server weights. We evaluate our approach on several computer vision and medical imaging datasets to demonstrate its effectiveness. To the best of our knowledge, this work is one of the first works in employing federated learning for segmentation, devoid of gradients or model information exchange. Code: https://github.com/JayParanjape/blackfed/tree/master
S-SAM: SVD-based Fine-Tuning of Segment Anything Model for Medical Image Segmentation
Aug 12, 2024Abstract:Medical image segmentation has been traditionally approached by training or fine-tuning the entire model to cater to any new modality or dataset. However, this approach often requires tuning a large number of parameters during training. With the introduction of the Segment Anything Model (SAM) for prompted segmentation of natural images, many efforts have been made towards adapting it efficiently for medical imaging, thus reducing the training time and resources. However, these methods still require expert annotations for every image in the form of point prompts or bounding box prompts during training and inference, making it tedious to employ them in practice. In this paper, we propose an adaptation technique, called S-SAM, that only trains parameters equal to 0.4% of SAM's parameters and at the same time uses simply the label names as prompts for producing precise masks. This not only makes tuning SAM more efficient than the existing adaptation methods but also removes the burden of providing expert prompts. We call this modified version S-SAM and evaluate it on five different modalities including endoscopic images, x-ray, ultrasound, CT, and histology images. Our experiments show that S-SAM outperforms state-of-the-art methods as well as existing SAM adaptation methods while tuning a significantly less number of parameters. We release the code for S-SAM at https://github.com/JayParanjape/SVDSAM.
Blackbox Adaptation for Medical Image Segmentation
May 17, 2024Abstract:In recent years, various large foundation models have been proposed for image segmentation. There models are often trained on large amounts of data corresponding to general computer vision tasks. Hence, these models do not perform well on medical data. There have been some attempts in the literature to perform parameter-efficient finetuning of such foundation models for medical image segmentation. However, these approaches assume that all the parameters of the model are available for adaptation. But, in many cases, these models are released as APIs or blackboxes, with no or limited access to the model parameters and data. In addition, finetuning methods also require a significant amount of compute, which may not be available for the downstream task. At the same time, medical data can't be shared with third-party agents for finetuning due to privacy reasons. To tackle these challenges, we pioneer a blackbox adaptation technique for prompted medical image segmentation, called BAPS. BAPS has two components - (i) An Image-Prompt decoder (IP decoder) module that generates visual prompts given an image and a prompt, and (ii) A Zero Order Optimization (ZOO) Method, called SPSA-GC that is used to update the IP decoder without the need for backpropagating through the foundation model. Thus, our method does not require any knowledge about the foundation model's weights or gradients. We test BAPS on four different modalities and show that our method can improve the original model's performance by around 4%.
An Endoscopic Chisel: Intraoperative Imaging Carves 3D Anatomical Models
Feb 19, 2024Abstract:Purpose: Preoperative imaging plays a pivotal role in sinus surgery where CTs offer patient-specific insights of complex anatomy, enabling real-time intraoperative navigation to complement endoscopy imaging. However, surgery elicits anatomical changes not represented in the preoperative model, generating an inaccurate basis for navigation during surgery progression. Methods: We propose a first vision-based approach to update the preoperative 3D anatomical model leveraging intraoperative endoscopic video for navigated sinus surgery where relative camera poses are known. We rely on comparisons of intraoperative monocular depth estimates and preoperative depth renders to identify modified regions. The new depths are integrated in these regions through volumetric fusion in a truncated signed distance function representation to generate an intraoperative 3D model that reflects tissue manipulation. Results: We quantitatively evaluate our approach by sequentially updating models for a five-step surgical progression in an ex vivo specimen. We compute the error between correspondences from the updated model and ground-truth intraoperative CT in the region of anatomical modification. The resulting models show a decrease in error during surgical progression as opposed to increasing when no update is employed. Conclusion: Our findings suggest that preoperative 3D anatomical models can be updated using intraoperative endoscopy video in navigated sinus surgery. Future work will investigate improvements to monocular depth estimation as well as removing the need for external navigation systems. The resulting ability to continuously update the patient model may provide surgeons with a more precise understanding of the current anatomical state and paves the way toward a digital twin paradigm for sinus surgery.
A Quantitative Evaluation of Dense 3D Reconstruction of Sinus Anatomy from Monocular Endoscopic Video
Oct 22, 2023



Abstract:Generating accurate 3D reconstructions from endoscopic video is a promising avenue for longitudinal radiation-free analysis of sinus anatomy and surgical outcomes. Several methods for monocular reconstruction have been proposed, yielding visually pleasant 3D anatomical structures by retrieving relative camera poses with structure-from-motion-type algorithms and fusion of monocular depth estimates. However, due to the complex properties of the underlying algorithms and endoscopic scenes, the reconstruction pipeline may perform poorly or fail unexpectedly. Further, acquiring medical data conveys additional challenges, presenting difficulties in quantitatively benchmarking these models, understanding failure cases, and identifying critical components that contribute to their precision. In this work, we perform a quantitative analysis of a self-supervised approach for sinus reconstruction using endoscopic sequences paired with optical tracking and high-resolution computed tomography acquired from nine ex-vivo specimens. Our results show that the generated reconstructions are in high agreement with the anatomy, yielding an average point-to-mesh error of 0.91 mm between reconstructions and CT segmentations. However, in a point-to-point matching scenario, relevant for endoscope tracking and navigation, we found average target registration errors of 6.58 mm. We identified that pose and depth estimation inaccuracies contribute equally to this error and that locally consistent sequences with shorter trajectories generate more accurate reconstructions. These results suggest that achieving global consistency between relative camera poses and estimated depths with the anatomy is essential. In doing so, we can ensure proper synergy between all components of the pipeline for improved reconstructions that will facilitate clinical application of this innovative technology.
AdaptiveSAM: Towards Efficient Tuning of SAM for Surgical Scene Segmentation
Aug 07, 2023Abstract:Segmentation is a fundamental problem in surgical scene analysis using artificial intelligence. However, the inherent data scarcity in this domain makes it challenging to adapt traditional segmentation techniques for this task. To tackle this issue, current research employs pretrained models and finetunes them on the given data. Even so, these require training deep networks with millions of parameters every time new data becomes available. A recently published foundation model, Segment-Anything (SAM), generalizes well to a large variety of natural images, hence tackling this challenge to a reasonable extent. However, SAM does not generalize well to the medical domain as is without utilizing a large amount of compute resources for fine-tuning and using task-specific prompts. Moreover, these prompts are in the form of bounding-boxes or foreground/background points that need to be annotated explicitly for every image, making this solution increasingly tedious with higher data size. In this work, we propose AdaptiveSAM - an adaptive modification of SAM that can adjust to new datasets quickly and efficiently, while enabling text-prompted segmentation. For finetuning AdaptiveSAM, we propose an approach called bias-tuning that requires a significantly smaller number of trainable parameters than SAM (less than 2\%). At the same time, AdaptiveSAM requires negligible expert intervention since it uses free-form text as prompt and can segment the object of interest with just the label name as prompt. Our experiments show that AdaptiveSAM outperforms current state-of-the-art methods on various medical imaging datasets including surgery, ultrasound and X-ray. Code is available at https://github.com/JayParanjape/biastuning
Cross-Dataset Adaptation for Instrument Classification in Cataract Surgery Videos
Jul 31, 2023Abstract:Surgical tool presence detection is an important part of the intra-operative and post-operative analysis of a surgery. State-of-the-art models, which perform this task well on a particular dataset, however, perform poorly when tested on another dataset. This occurs due to a significant domain shift between the datasets resulting from the use of different tools, sensors, data resolution etc. In this paper, we highlight this domain shift in the commonly performed cataract surgery and propose a novel end-to-end Unsupervised Domain Adaptation (UDA) method called the Barlow Adaptor that addresses the problem of distribution shift without requiring any labels from another domain. In addition, we introduce a novel loss called the Barlow Feature Alignment Loss (BFAL) which aligns features across different domains while reducing redundancy and the need for higher batch sizes, thus improving cross-dataset performance. The use of BFAL is a novel approach to address the challenge of domain shift in cataract surgery data. Extensive experiments are conducted on two cataract surgery datasets and it is shown that the proposed method outperforms the state-of-the-art UDA methods by 6%. The code can be found at https://github.com/JayParanjape/Barlow-Adaptor
GLSFormer: Gated - Long, Short Sequence Transformer for Step Recognition in Surgical Videos
Jul 20, 2023Abstract:Automated surgical step recognition is an important task that can significantly improve patient safety and decision-making during surgeries. Existing state-of-the-art methods for surgical step recognition either rely on separate, multi-stage modeling of spatial and temporal information or operate on short-range temporal resolution when learned jointly. However, the benefits of joint modeling of spatio-temporal features and long-range information are not taken in account. In this paper, we propose a vision transformer-based approach to jointly learn spatio-temporal features directly from sequence of frame-level patches. Our method incorporates a gated-temporal attention mechanism that intelligently combines short-term and long-term spatio-temporal feature representations. We extensively evaluate our approach on two cataract surgery video datasets, namely Cataract-101 and D99, and demonstrate superior performance compared to various state-of-the-art methods. These results validate the suitability of our proposed approach for automated surgical step recognition. Our code is released at: https://github.com/nisargshah1999/GLSFormer
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge