Ben Walker
Combining Hough Transform and Deep Learning Approaches to Reconstruct ECG Signals From Printouts
Oct 18, 2024Abstract:This work presents our team's (SignalSavants) winning contribution to the 2024 George B. Moody PhysioNet Challenge. The Challenge had two goals: reconstruct ECG signals from printouts and classify them for cardiac diseases. Our focus was the first task. Despite many ECGs being digitally recorded today, paper ECGs remain common throughout the world. Digitising them could help build more diverse datasets and enable automated analyses. However, the presence of varying recording standards and poor image quality requires a data-centric approach for developing robust models that can generalise effectively. Our approach combines the creation of a diverse training set, Hough transform to rotate images, a U-Net based segmentation model to identify individual signals, and mask vectorisation to reconstruct the signals. We assessed the performance of our models using the 10-fold stratified cross-validation (CV) split of 21,799 recordings proposed by the PTB-XL dataset. On the digitisation task, our model achieved an average CV signal-to-noise ratio of 17.02 and an official Challenge score of 12.15 on the hidden set, securing first place in the competition. Our study shows the challenges of building robust, generalisable, digitisation approaches. Such models require large amounts of resources (data, time, and computational power) but have great potential in diversifying the data available.
Multimodal deep learning approach to predicting neurological recovery from coma after cardiac arrest
Mar 09, 2024



Abstract:This work showcases our team's (The BEEGees) contributions to the 2023 George B. Moody PhysioNet Challenge. The aim was to predict neurological recovery from coma following cardiac arrest using clinical data and time-series such as multi-channel EEG and ECG signals. Our modelling approach is multimodal, based on two-dimensional spectrogram representations derived from numerous EEG channels, alongside the integration of clinical data and features extracted directly from EEG recordings. Our submitted model achieved a Challenge score of $0.53$ on the hidden test set for predictions made $72$ hours after return of spontaneous circulation. Our study shows the efficacy and limitations of employing transfer learning in medical classification. With regard to prospective implementation, our analysis reveals that the performance of the model is strongly linked to the selection of a decision threshold and exhibits strong variability across data splits.
Unbiased Decisions Reduce Regret: Adversarial Domain Adaptation for the Bank Loan Problem
Aug 15, 2023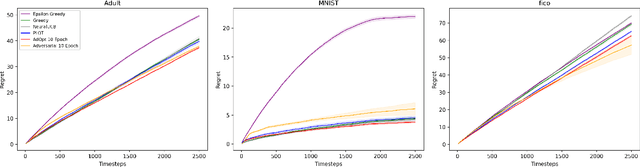
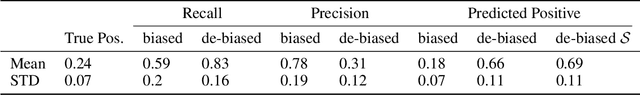
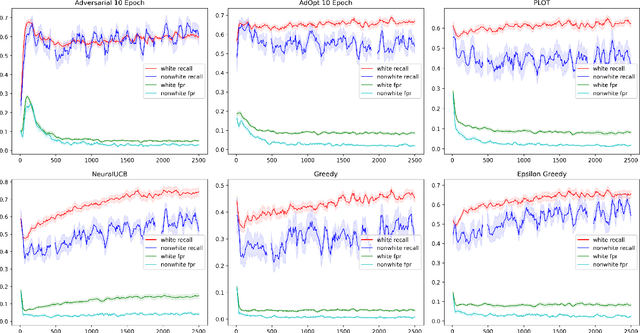

Abstract:In many real world settings binary classification decisions are made based on limited data in near real-time, e.g. when assessing a loan application. We focus on a class of these problems that share a common feature: the true label is only observed when a data point is assigned a positive label by the principal, e.g. we only find out whether an applicant defaults if we accepted their loan application. As a consequence, the false rejections become self-reinforcing and cause the labelled training set, that is being continuously updated by the model decisions, to accumulate bias. Prior work mitigates this effect by injecting optimism into the model, however this comes at the cost of increased false acceptance rate. We introduce adversarial optimism (AdOpt) to directly address bias in the training set using adversarial domain adaptation. The goal of AdOpt is to learn an unbiased but informative representation of past data, by reducing the distributional shift between the set of accepted data points and all data points seen thus far. AdOpt significantly exceeds state-of-the-art performance on a set of challenging benchmark problems. Our experiments also provide initial evidence that the introduction of adversarial domain adaptation improves fairness in this setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge