Burooj Ghani
Decodable but not structured: linear probing enables Underwater Acoustic Target Recognition with pretrained audio embeddings
Jan 13, 2026Abstract:Increasing levels of anthropogenic noise from ships contribute significantly to underwater sound pollution, posing risks to marine ecosystems. This makes monitoring crucial to understand and quantify the impact of the ship radiated noise. Passive Acoustic Monitoring (PAM) systems are widely deployed for this purpose, generating years of underwater recordings across diverse soundscapes. Manual analysis of such large-scale data is impractical, motivating the need for automated approaches based on machine learning. Recent advances in automatic Underwater Acoustic Target Recognition (UATR) have largely relied on supervised learning, which is constrained by the scarcity of labeled data. Transfer Learning (TL) offers a promising alternative to mitigate this limitation. In this work, we conduct the first empirical comparative study of transfer learning for UATR, evaluating multiple pretrained audio models originating from diverse audio domains. The pretrained model weights are frozen, and the resulting embeddings are analyzed through classification, clustering, and similarity-based evaluations. The analysis shows that the geometrical structure of the embedding space is largely dominated by recording-specific characteristics. However, a simple linear probe can effectively suppress this recording-specific information and isolate ship-type features from these embeddings. As a result, linear probing enables effective automatic UATR using pretrained audio models at low computational cost, significantly reducing the need for a large amounts of high-quality labeled ship recordings.
Automated data curation for self-supervised learning in underwater acoustic analysis
May 26, 2025Abstract:The sustainability of the ocean ecosystem is threatened by increased levels of sound pollution, making monitoring crucial to understand its variability and impact. Passive acoustic monitoring (PAM) systems collect a large amount of underwater sound recordings, but the large volume of data makes manual analysis impossible, creating the need for automation. Although machine learning offers a potential solution, most underwater acoustic recordings are unlabeled. Self-supervised learning models have demonstrated success in learning from large-scale unlabeled data in various domains like computer vision, Natural Language Processing, and audio. However, these models require large, diverse, and balanced datasets for training in order to generalize well. To address this, a fully automated self-supervised data curation pipeline is proposed to create a diverse and balanced dataset from raw PAM data. It integrates Automatic Identification System (AIS) data with recordings from various hydrophones in the U.S. waters. Using hierarchical k-means clustering, the raw audio data is sampled and then combined with AIS samples to create a balanced and diverse dataset. The resulting curated dataset enables the development of self-supervised learning models, facilitating various tasks such as monitoring marine mammals and assessing sound pollution.
Clustering and novel class recognition: evaluating bioacoustic deep learning feature extractors
Apr 09, 2025



Abstract:In computational bioacoustics, deep learning models are composed of feature extractors and classifiers. The feature extractors generate vector representations of the input sound segments, called embeddings, which can be input to a classifier. While benchmarking of classification scores provides insights into specific performance statistics, it is limited to species that are included in the models' training data. Furthermore, it makes it impossible to compare models trained on very different taxonomic groups. This paper aims to address this gap by analyzing the embeddings generated by the feature extractors of 15 bioacoustic models spanning a wide range of setups (model architectures, training data, training paradigms). We evaluated and compared different ways in which models structure embedding spaces through clustering and kNN classification, which allows us to focus our comparison on feature extractors independent of their classifiers. We believe that this approach lets us evaluate the adaptability and generalization potential of models going beyond the classes they were trained on.
InsectSet459: an open dataset of insect sounds for bioacoustic machine learning
Mar 19, 2025Abstract:Automatic recognition of insect sound could help us understand changing biodiversity trends around the world -- but insect sounds are challenging to recognize even for deep learning. We present a new dataset comprised of 26399 audio files, from 459 species of Orthoptera and Cicadidae. It is the first large-scale dataset of insect sound that is easily applicable for developing novel deep-learning methods. Its recordings were made with a variety of audio recorders using varying sample rates to capture the extremely broad range of frequencies that insects produce. We benchmark performance with two state-of-the-art deep learning classifiers, demonstrating good performance but also significant room for improvement in acoustic insect classification. This dataset can serve as a realistic test case for implementing insect monitoring workflows, and as a challenging basis for the development of audio representation methods that can handle highly variable frequencies and/or sample rates.
Generalization in birdsong classification: impact of transfer learning methods and dataset characteristics
Sep 21, 2024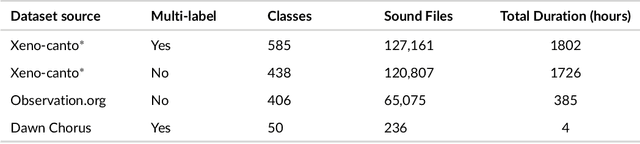
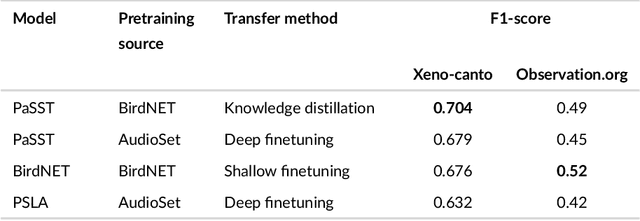

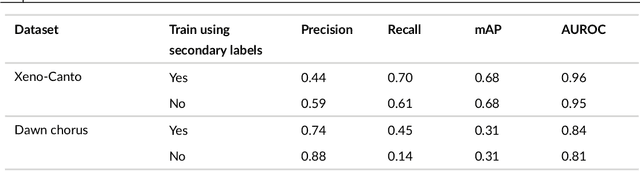
Abstract:Animal sounds can be recognised automatically by machine learning, and this has an important role to play in biodiversity monitoring. Yet despite increasingly impressive capabilities, bioacoustic species classifiers still exhibit imbalanced performance across species and habitats, especially in complex soundscapes. In this study, we explore the effectiveness of transfer learning in large-scale bird sound classification across various conditions, including single- and multi-label scenarios, and across different model architectures such as CNNs and Transformers. Our experiments demonstrate that both fine-tuning and knowledge distillation yield strong performance, with cross-distillation proving particularly effective in improving in-domain performance on Xeno-canto data. However, when generalizing to soundscapes, shallow fine-tuning exhibits superior performance compared to knowledge distillation, highlighting its robustness and constrained nature. Our study further investigates how to use multi-species labels, in cases where these are present but incomplete. We advocate for more comprehensive labeling practices within the animal sound community, including annotating background species and providing temporal details, to enhance the training of robust bird sound classifiers. These findings provide insights into the optimal reuse of pretrained models for advancing automatic bioacoustic recognition.
Mind the Domain Gap: a Systematic Analysis on Bioacoustic Sound Event Detection
Mar 27, 2024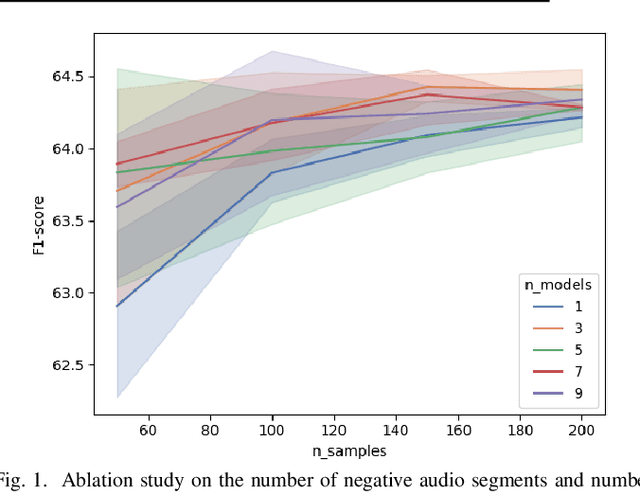
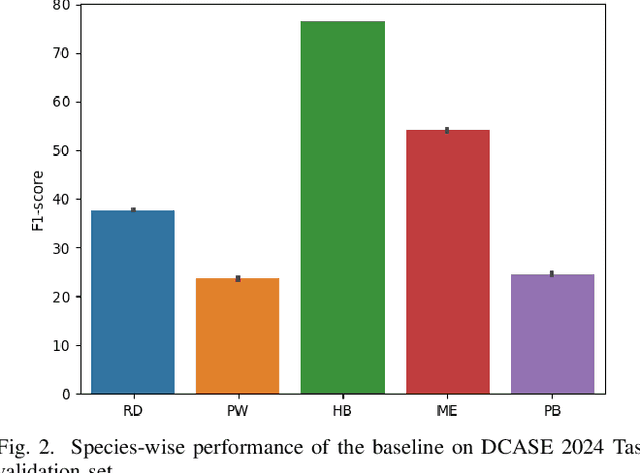
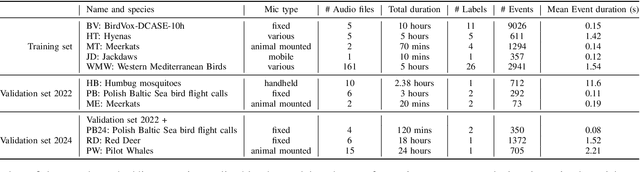
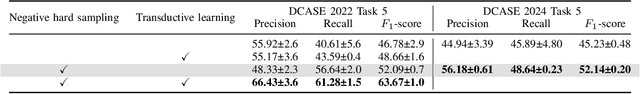
Abstract:Detecting the presence of animal vocalisations in nature is essential to study animal populations and their behaviors. A recent development in the field is the introduction of the task known as few-shot bioacoustic sound event detection, which aims to train a versatile animal sound detector using only a small set of audio samples. Previous efforts in this area have utilized different architectures and data augmentation techniques to enhance model performance. However, these approaches have not fully bridged the domain gap between source and target distributions, limiting their applicability in real-world scenarios. In this work, we introduce an new dataset designed to augment the diversity and breadth of classes available for few-shot bioacoustic event detection, building on the foundations of our previous datasets. To establish a robust baseline system tailored for the DCASE 2024 Task 5 challenge, we delve into an array of acoustic features and adopt negative hard sampling as our primary domain adaptation strategy. This approach, chosen in alignment with the challenge's guidelines that necessitate the independent treatment of each audio file, sidesteps the use of transductive learning to ensure compliance while aiming to enhance the system's adaptability to domain shifts. Our experiments show that the proposed baseline system achieves a better performance compared with the vanilla prototypical network. The findings also confirm the effectiveness of each domain adaptation method by ablating different components within the networks. This highlights the potential to improve few-shot bioacoustic sound event detection by further reducing the impact of domain shift.
Efficient speech detection in environmental audio using acoustic recognition and knowledge distillation
Dec 14, 2023


Abstract:The ongoing biodiversity crisis, driven by factors such as land-use change and global warming, emphasizes the need for effective ecological monitoring methods. Acoustic monitoring of biodiversity has emerged as an important monitoring tool. Detecting human voices in soundscape monitoring projects is useful both for analysing human disturbance and for privacy filtering. Despite significant strides in deep learning in recent years, the deployment of large neural networks on compact devices poses challenges due to memory and latency constraints. Our approach focuses on leveraging knowledge distillation techniques to design efficient, lightweight student models for speech detection in bioacoustics. In particular, we employed the MobileNetV3-Small-Pi model to create compact yet effective student architectures to compare against the larger EcoVAD teacher model, a well-regarded voice detection architecture in eco-acoustic monitoring. The comparative analysis included examining various configurations of the MobileNetV3-Small-Pi derived student models to identify optimal performance. Additionally, a thorough evaluation of different distillation techniques was conducted to ascertain the most effective method for model selection. Our findings revealed that the distilled models exhibited comparable performance to the EcoVAD teacher model, indicating a promising approach to overcoming computational barriers for real-time ecological monitoring.
Feature Embeddings from Large-Scale Acoustic Bird Classifiers Enable Few-Shot Transfer Learning
Jul 12, 2023Abstract:Automated bioacoustic analysis aids understanding and protection of both marine and terrestrial animals and their habitats across extensive spatiotemporal scales, and typically involves analyzing vast collections of acoustic data. With the advent of deep learning models, classification of important signals from these datasets has markedly improved. These models power critical data analyses for research and decision-making in biodiversity monitoring, animal behaviour studies, and natural resource management. However, deep learning models are often data-hungry and require a significant amount of labeled training data to perform well. While sufficient training data is available for certain taxonomic groups (e.g., common bird species), many classes (such as rare and endangered species, many non-bird taxa, and call-type), lack enough data to train a robust model from scratch. This study investigates the utility of feature embeddings extracted from large-scale audio classification models to identify bioacoustic classes other than the ones these models were originally trained on. We evaluate models on diverse datasets, including different bird calls and dialect types, bat calls, marine mammals calls, and amphibians calls. The embeddings extracted from the models trained on bird vocalization data consistently allowed higher quality classification than the embeddings trained on general audio datasets. The results of this study indicate that high-quality feature embeddings from large-scale acoustic bird classifiers can be harnessed for few-shot transfer learning, enabling the learning of new classes from a limited quantity of training data. Our findings reveal the potential for efficient analyses of novel bioacoustic tasks, even in scenarios where available training data is limited to a few samples.
Few-shot bioacoustic event detection at the DCASE 2023 challenge
Jun 15, 2023



Abstract:Few-shot bioacoustic event detection consists in detecting sound events of specified types, in varying soundscapes, while having access to only a few examples of the class of interest. This task ran as part of the DCASE challenge for the third time this year with an evaluation set expanded to include new animal species, and a new rule: ensemble models were no longer allowed. The 2023 few shot task received submissions from 6 different teams with F-scores reaching as high as 63% on the evaluation set. Here we describe the task, focusing on describing the elements that differed from previous years. We also take a look back at past editions to describe how the task has evolved. Not only have the F-score results steadily improved (40% to 60% to 63%), but the type of systems proposed have also become more complex. Sound event detection systems are no longer simple variations of the baselines provided: multiple few-shot learning methodologies are still strong contenders for the task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge