Vincent J. Kalkman
Generalization in birdsong classification: impact of transfer learning methods and dataset characteristics
Sep 21, 2024
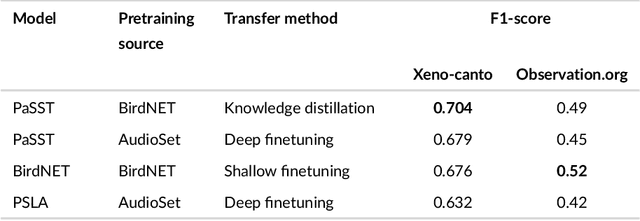

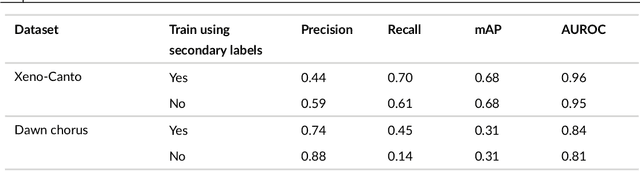
Abstract:Animal sounds can be recognised automatically by machine learning, and this has an important role to play in biodiversity monitoring. Yet despite increasingly impressive capabilities, bioacoustic species classifiers still exhibit imbalanced performance across species and habitats, especially in complex soundscapes. In this study, we explore the effectiveness of transfer learning in large-scale bird sound classification across various conditions, including single- and multi-label scenarios, and across different model architectures such as CNNs and Transformers. Our experiments demonstrate that both fine-tuning and knowledge distillation yield strong performance, with cross-distillation proving particularly effective in improving in-domain performance on Xeno-canto data. However, when generalizing to soundscapes, shallow fine-tuning exhibits superior performance compared to knowledge distillation, highlighting its robustness and constrained nature. Our study further investigates how to use multi-species labels, in cases where these are present but incomplete. We advocate for more comprehensive labeling practices within the animal sound community, including annotating background species and providing temporal details, to enhance the training of robust bird sound classifiers. These findings provide insights into the optimal reuse of pretrained models for advancing automatic bioacoustic recognition.
Performance of computer vision algorithms for fine-grained classification using crowdsourced insect images
Apr 04, 2024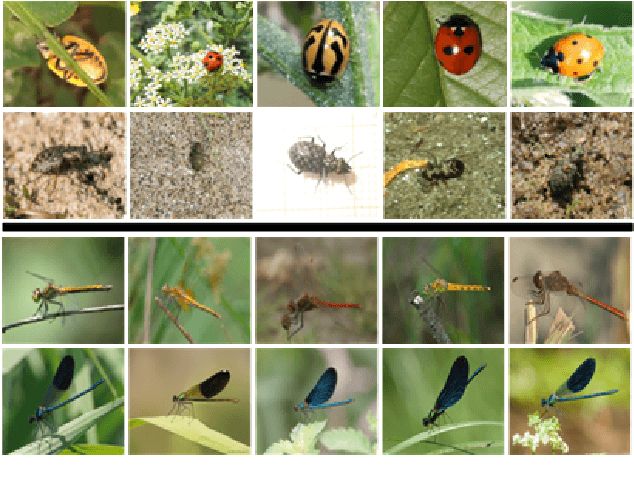
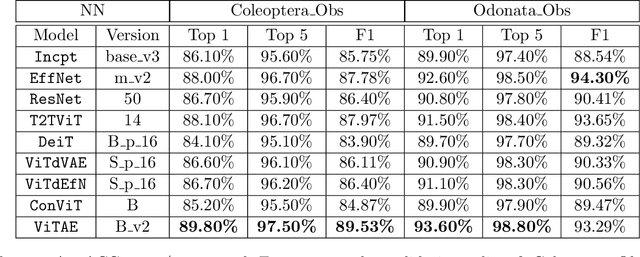

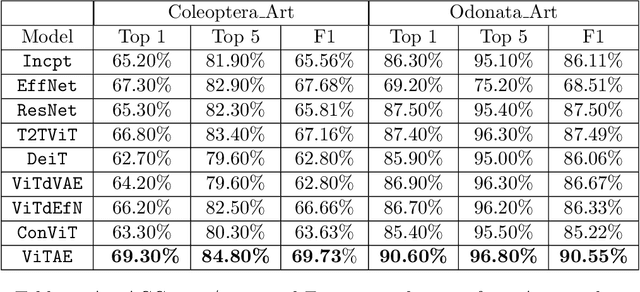
Abstract:With fine-grained classification, we identify unique characteristics to distinguish among classes of the same super-class. We are focusing on species recognition in Insecta, as they are critical for biodiversity monitoring and at the base of many ecosystems. With citizen science campaigns, billions of images are collected in the wild. Once these are labelled, experts can use them to create distribution maps. However, the labelling process is time-consuming, which is where computer vision comes in. The field of computer vision offers a wide range of algorithms, each with its strengths and weaknesses; how do we identify the algorithm that is in line with our application? To answer this question, we provide a full and detailed evaluation of nine algorithms among deep convolutional networks (CNN), vision transformers (ViT), and locality-based vision transformers (LBVT) on 4 different aspects: classification performance, embedding quality, computational cost, and gradient activity. We offer insights that we haven't yet had in this domain proving to which extent these algorithms solve the fine-grained tasks in Insecta. We found that the ViT performs the best on inference speed and computational cost while the LBVT outperforms the others on performance and embedding quality; the CNN provide a trade-off among the metrics.
Comparison between transformers and convolutional models for fine-grained classification of insects
Jul 20, 2023Abstract:Fine-grained classification is challenging due to the difficulty of finding discriminatory features. This problem is exacerbated when applied to identifying species within the same taxonomical class. This is because species are often sharing morphological characteristics that make them difficult to differentiate. We consider the taxonomical class of Insecta. The identification of insects is essential in biodiversity monitoring as they are one of the inhabitants at the base of many ecosystems. Citizen science is doing brilliant work of collecting images of insects in the wild giving the possibility to experts to create improved distribution maps in all countries. We have billions of images that need to be automatically classified and deep neural network algorithms are one of the main techniques explored for fine-grained tasks. At the SOTA, the field of deep learning algorithms is extremely fruitful, so how to identify the algorithm to use? We focus on Odonata and Coleoptera orders, and we propose an initial comparative study to analyse the two best-known layer structures for computer vision: transformer and convolutional layers. We compare the performance of T2TViT, a fully transformer-base, EfficientNet, a fully convolutional-base, and ViTAE, a hybrid. We analyse the performance of the three models in identical conditions evaluating the performance per species, per morph together with sex, the inference time, and the overall performance with unbalanced datasets of images from smartphones. Although we observe high performances with all three families of models, our analysis shows that the hybrid model outperforms the fully convolutional-base and fully transformer-base models on accuracy performance and the fully transformer-base model outperforms the others on inference speed and, these prove the transformer to be robust to the shortage of samples and to be faster at inference time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge