Eli M Carrami
Season combinatorial intervention predictions with Salt & Peper
Apr 25, 2024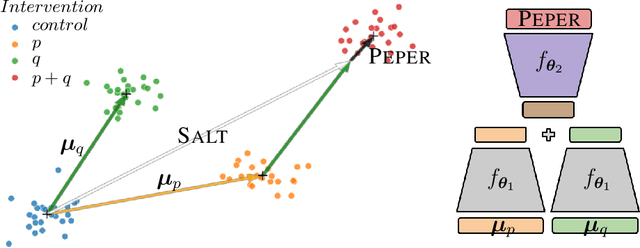
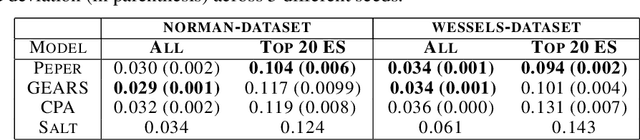
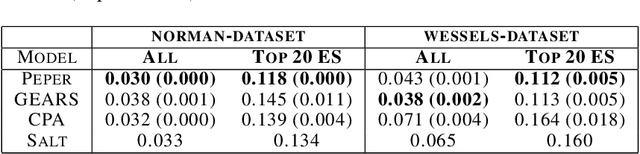
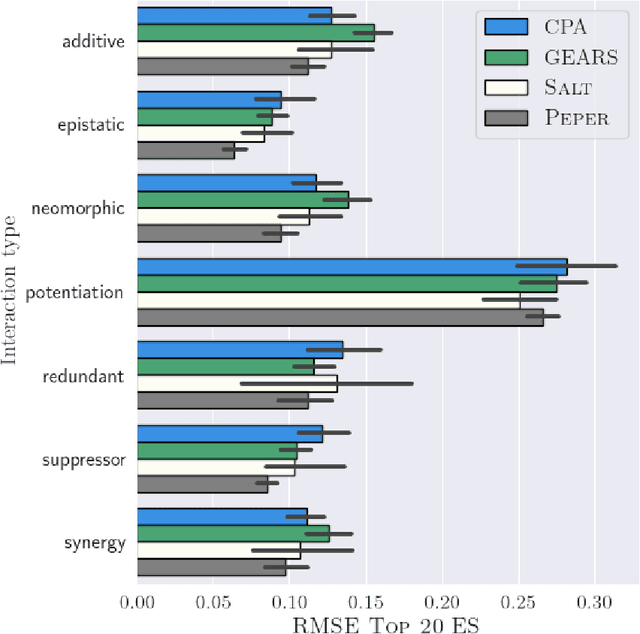
Abstract:Interventions play a pivotal role in the study of complex biological systems. In drug discovery, genetic interventions (such as CRISPR base editing) have become central to both identifying potential therapeutic targets and understanding a drug's mechanism of action. With the advancement of CRISPR and the proliferation of genome-scale analyses such as transcriptomics, a new challenge is to navigate the vast combinatorial space of concurrent genetic interventions. Addressing this, our work concentrates on estimating the effects of pairwise genetic combinations on the cellular transcriptome. We introduce two novel contributions: Salt, a biologically-inspired baseline that posits the mostly additive nature of combination effects, and Peper, a deep learning model that extends Salt's additive assumption to achieve unprecedented accuracy. Our comprehensive comparison against existing state-of-the-art methods, grounded in diverse metrics, and our out-of-distribution analysis highlight the limitations of current models in realistic settings. This analysis underscores the necessity for improved modelling techniques and data acquisition strategies, paving the way for more effective exploration of genetic intervention effects.
PQA: Zero-shot Protein Question Answering for Free-form Scientific Enquiry with Large Language Models
Feb 21, 2024



Abstract:We introduce the novel task of zero-shot Protein Question Answering (PQA) for free-form scientific enquiry. Given a previously unseen protein sequence and a natural language question, the task is to deliver a scientifically accurate answer. This task not only supports future biological research, but could also provide a test bed for assessing the scientific precision of large language models (LLMs). We contribute the first specialized dataset for PQA model training, containing 257K protein sequences annotated with 1.97M scientific question-answer pairs. Additionally, we propose and study several novel biologically relevant benchmarks for scientific PQA. Employing two robust multi-modal architectures, we establish an initial state-of-the-art performance for PQA and reveal key performance factors through ablation studies. Our comprehensive PQA framework, named Pika, including dataset, code, model checkpoints, and a user-friendly demo, is openly accessible on github.com/EMCarrami/Pika, promoting wider research and application in the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge