Generalizability of Machine Learning Models: Quantitative Evaluation of Three Methodological Pitfalls
Paper and Code
Feb 01, 2022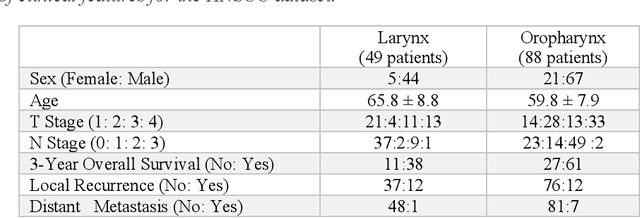
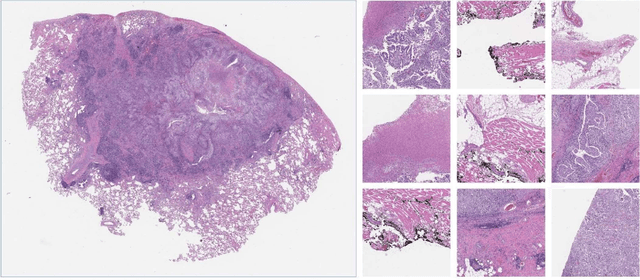
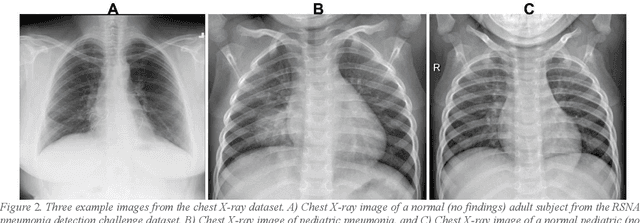
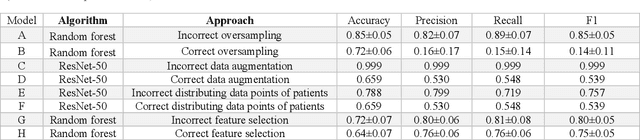
Despite the great potential of machine learning, the lack of generalizability has hindered the widespread adoption of these technologies in routine clinical practice. We investigate three methodological pitfalls: (1) violation of independence assumption, (2) model evaluation with an inappropriate performance indicator, and (3) batch effect and how these pitfalls could affect the generalizability of machine learning models. We implement random forest and deep convolutional neural network models using several medical imaging datasets, including head and neck CT, lung CT, chest X-Ray, and histopathological images, to quantify and illustrate the effect of these pitfalls. We develop these models with and without the pitfall and compare the performance of the resulting models in terms of accuracy, precision, recall, and F1 score. Our results showed that violation of the independence assumption could substantially affect model generalizability. More specifically, (I) applying oversampling before splitting data into train, validation and test sets; (II) performing data augmentation before splitting data; (III) distributing data points for a subject across training, validation, and test sets; and (IV) applying feature selection before splitting data led to superficial boosts in model performance. We also observed that inappropriate performance indicators could lead to erroneous conclusions. Also, batch effect could lead to developing models that lack generalizability. The aforementioned methodological pitfalls lead to machine learning models with over-optimistic performance. These errors, if made, cannot be captured using internal model evaluation, and the inaccurate predictions made by the model may lead to wrong conclusions and interpretations. Therefore, avoiding these pitfalls is a necessary condition for developing generalizable models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge