Jayashree Kalpathy-Cramer
The Massachusetts General Hospital, USA and University of Colorado, USA
Fair Evaluation of Federated Learning Algorithms for Automated Breast Density Classification: The Results of the 2022 ACR-NCI-NVIDIA Federated Learning Challenge
May 22, 2024Abstract:The correct interpretation of breast density is important in the assessment of breast cancer risk. AI has been shown capable of accurately predicting breast density, however, due to the differences in imaging characteristics across mammography systems, models built using data from one system do not generalize well to other systems. Though federated learning (FL) has emerged as a way to improve the generalizability of AI without the need to share data, the best way to preserve features from all training data during FL is an active area of research. To explore FL methodology, the breast density classification FL challenge was hosted in partnership with the American College of Radiology, Harvard Medical School's Mass General Brigham, University of Colorado, NVIDIA, and the National Institutes of Health National Cancer Institute. Challenge participants were able to submit docker containers capable of implementing FL on three simulated medical facilities, each containing a unique large mammography dataset. The breast density FL challenge ran from June 15 to September 5, 2022, attracting seven finalists from around the world. The winning FL submission reached a linear kappa score of 0.653 on the challenge test data and 0.413 on an external testing dataset, scoring comparably to a model trained on the same data in a central location.
* 16 pages, 9 figures
Deep Learning-based Prediction of Breast Cancer Tumor and Immune Phenotypes from Histopathology
Apr 25, 2024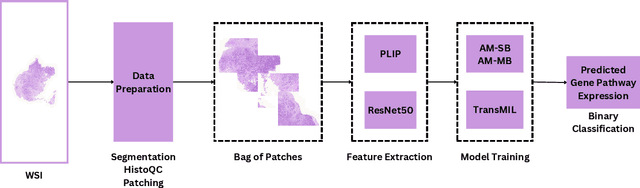

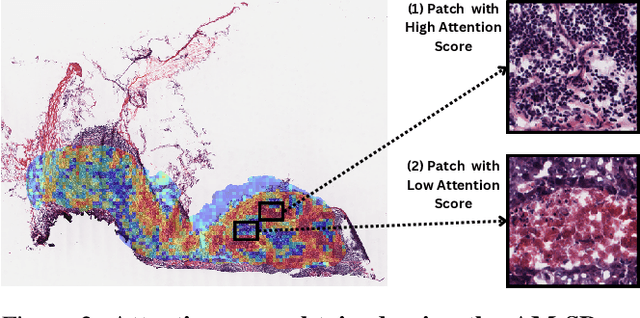

Abstract:The interactions between tumor cells and the tumor microenvironment (TME) dictate therapeutic efficacy of radiation and many systemic therapies in breast cancer. However, to date, there is not a widely available method to reproducibly measure tumor and immune phenotypes for each patient's tumor. Given this unmet clinical need, we applied multiple instance learning (MIL) algorithms to assess activity of ten biologically relevant pathways from the hematoxylin and eosin (H&E) slide of primary breast tumors. We employed different feature extraction approaches and state-of-the-art model architectures. Using binary classification, our models attained area under the receiver operating characteristic (AUROC) scores above 0.70 for nearly all gene expression pathways and on some cases, exceeded 0.80. Attention maps suggest that our trained models recognize biologically relevant spatial patterns of cell sub-populations from H&E. These efforts represent a first step towards developing computational H&E biomarkers that reflect facets of the TME and hold promise for augmenting precision oncology.
Patient-specific, mechanistic models of tumor growth incorporating artificial intelligence and big data
Aug 28, 2023Abstract:Despite the remarkable advances in cancer diagnosis, treatment, and management that have occurred over the past decade, malignant tumors remain a major public health problem. Further progress in combating cancer may be enabled by personalizing the delivery of therapies according to the predicted response for each individual patient. The design of personalized therapies requires patient-specific information integrated into an appropriate mathematical model of tumor response. A fundamental barrier to realizing this paradigm is the current lack of a rigorous, yet practical, mathematical theory of tumor initiation, development, invasion, and response to therapy. In this review, we begin by providing an overview of different approaches to modeling tumor growth and treatment, including mechanistic as well as data-driven models based on ``big data" and artificial intelligence. Next, we present illustrative examples of mathematical models manifesting their utility and discussing the limitations of stand-alone mechanistic and data-driven models. We further discuss the potential of mechanistic models for not only predicting, but also optimizing response to therapy on a patient-specific basis. We then discuss current efforts and future possibilities to integrate mechanistic and data-driven models. We conclude by proposing five fundamental challenges that must be addressed to fully realize personalized care for cancer patients driven by computational models.
A generalized framework to predict continuous scores from medical ordinal labels
May 30, 2023Abstract:Many variables of interest in clinical medicine, like disease severity, are recorded using discrete ordinal categories such as normal/mild/moderate/severe. These labels are used to train and evaluate disease severity prediction models. However, ordinal categories represent a simplification of an underlying continuous severity spectrum. Using continuous scores instead of ordinal categories is more sensitive to detecting small changes in disease severity over time. Here, we present a generalized framework that accurately predicts continuously valued variables using only discrete ordinal labels during model development. We found that for three clinical prediction tasks, models that take the ordinal relationship of the training labels into account outperformed conventional multi-class classification models. Particularly the continuous scores generated by ordinal classification and regression models showed a significantly higher correlation with expert rankings of disease severity and lower mean squared errors compared to the multi-class classification models. Furthermore, the use of MC dropout significantly improved the ability of all evaluated deep learning approaches to predict continuously valued scores that truthfully reflect the underlying continuous target variable. We showed that accurate continuously valued predictions can be generated even if the model development only involves discrete ordinal labels. The novel framework has been validated on three different clinical prediction tasks and has proven to bridge the gap between discrete ordinal labels and the underlying continuously valued variables.
MONAI: An open-source framework for deep learning in healthcare
Nov 04, 2022



Abstract:Artificial Intelligence (AI) is having a tremendous impact across most areas of science. Applications of AI in healthcare have the potential to improve our ability to detect, diagnose, prognose, and intervene on human disease. For AI models to be used clinically, they need to be made safe, reproducible and robust, and the underlying software framework must be aware of the particularities (e.g. geometry, physiology, physics) of medical data being processed. This work introduces MONAI, a freely available, community-supported, and consortium-led PyTorch-based framework for deep learning in healthcare. MONAI extends PyTorch to support medical data, with a particular focus on imaging, and provide purpose-specific AI model architectures, transformations and utilities that streamline the development and deployment of medical AI models. MONAI follows best practices for software-development, providing an easy-to-use, robust, well-documented, and well-tested software framework. MONAI preserves the simple, additive, and compositional approach of its underlying PyTorch libraries. MONAI is being used by and receiving contributions from research, clinical and industrial teams from around the world, who are pursuing applications spanning nearly every aspect of healthcare.
Towards More Efficient Data Valuation in Healthcare Federated Learning using Ensembling
Sep 12, 2022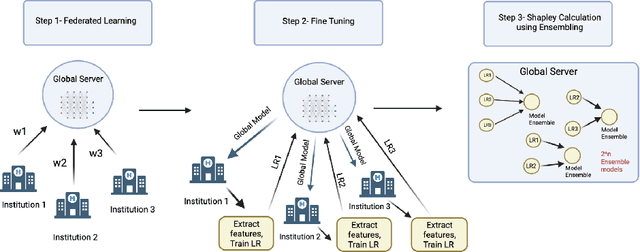
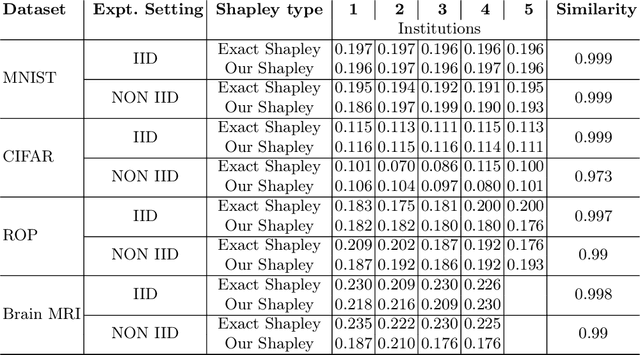

Abstract:Federated Learning (FL) wherein multiple institutions collaboratively train a machine learning model without sharing data is becoming popular. Participating institutions might not contribute equally, some contribute more data, some better quality data or some more diverse data. To fairly rank the contribution of different institutions, Shapley value (SV) has emerged as the method of choice. Exact SV computation is impossibly expensive, especially when there are hundreds of contributors. Existing SV computation techniques use approximations. However, in healthcare where the number of contributing institutions are likely not of a colossal scale, computing exact SVs is still exorbitantly expensive, but not impossible. For such settings, we propose an efficient SV computation technique called SaFE (Shapley Value for Federated Learning using Ensembling). We empirically show that SaFE computes values that are close to exact SVs, and that it performs better than current SV approximations. This is particularly relevant in medical imaging setting where widespread heterogeneity across institutions is rampant and fast accurate data valuation is required to determine the contribution of each participant in multi-institutional collaborative learning.
Estimating Test Performance for AI Medical Devices under Distribution Shift with Conformal Prediction
Jul 12, 2022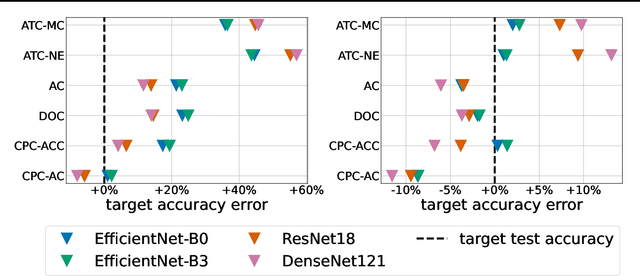
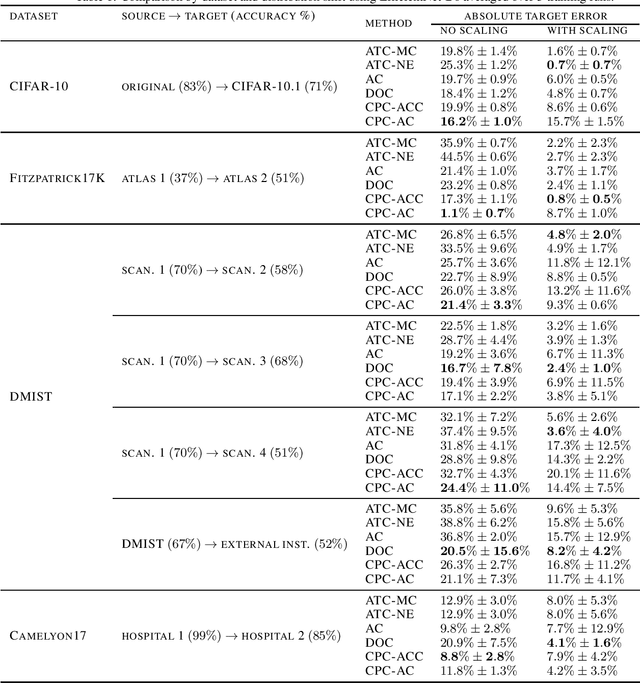
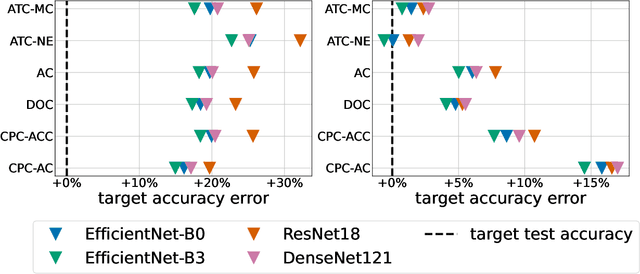
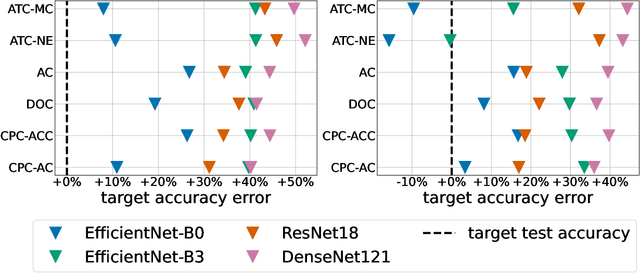
Abstract:Estimating the test performance of software AI-based medical devices under distribution shifts is crucial for evaluating the safety, efficiency, and usability prior to clinical deployment. Due to the nature of regulated medical device software and the difficulty in acquiring large amounts of labeled medical datasets, we consider the task of predicting the test accuracy of an arbitrary black-box model on an unlabeled target domain without modification to the original training process or any distributional assumptions of the original source data (i.e. we treat the model as a "black-box" and only use the predicted output responses). We propose a "black-box" test estimation technique based on conformal prediction and evaluate it against other methods on three medical imaging datasets (mammography, dermatology, and histopathology) under several clinically relevant types of distribution shift (institution, hardware scanner, atlas, hospital). We hope that by promoting practical and effective estimation techniques for black-box models, manufacturers of medical devices will develop more standardized and realistic evaluation procedures to improve the robustness and trustworthiness of clinical AI tools.
Three Applications of Conformal Prediction for Rating Breast Density in Mammography
Jun 23, 2022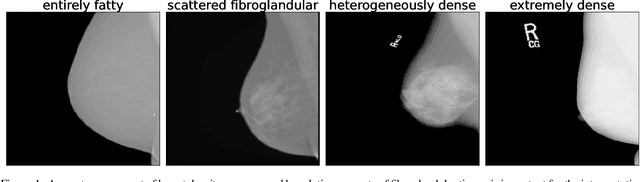
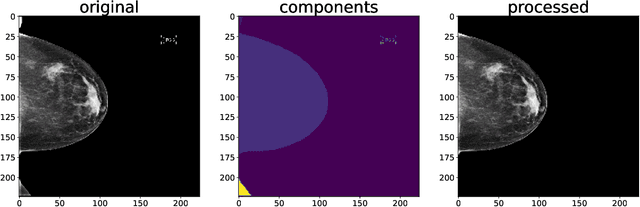
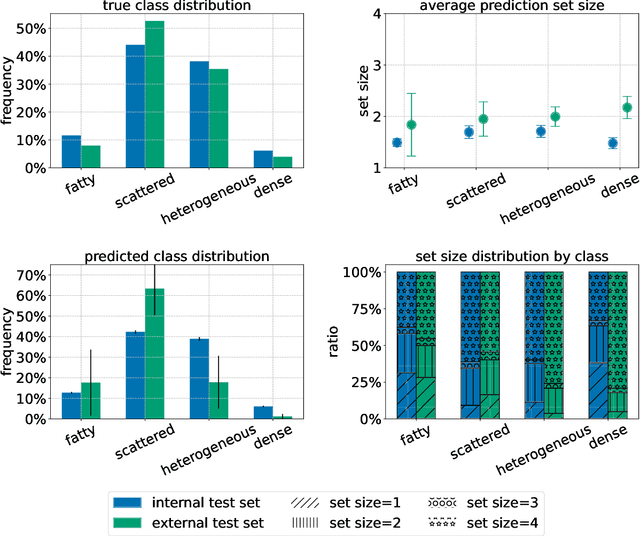
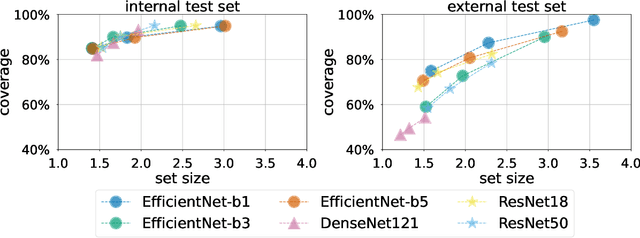
Abstract:Breast cancer is the most common cancers and early detection from mammography screening is crucial in improving patient outcomes. Assessing mammographic breast density is clinically important as the denser breasts have higher risk and are more likely to occlude tumors. Manual assessment by experts is both time-consuming and subject to inter-rater variability. As such, there has been increased interest in the development of deep learning methods for mammographic breast density assessment. Despite deep learning having demonstrated impressive performance in several prediction tasks for applications in mammography, clinical deployment of deep learning systems in still relatively rare; historically, mammography Computer-Aided Diagnoses (CAD) have over-promised and failed to deliver. This is in part due to the inability to intuitively quantify uncertainty of the algorithm for the clinician, which would greatly enhance usability. Conformal prediction is well suited to increase reliably and trust in deep learning tools but they lack realistic evaluations on medical datasets. In this paper, we present a detailed analysis of three possible applications of conformal prediction applied to medical imaging tasks: distribution shift characterization, prediction quality improvement, and subgroup fairness analysis. Our results show the potential of distribution-free uncertainty quantification techniques to enhance trust on AI algorithms and expedite their translation to usage.
Improving the repeatability of deep learning models with Monte Carlo dropout
Feb 15, 2022



Abstract:The integration of artificial intelligence into clinical workflows requires reliable and robust models. Repeatability is a key attribute of model robustness. Repeatable models output predictions with low variation during independent tests carried out under similar conditions. During model development and evaluation, much attention is given to classification performance while model repeatability is rarely assessed, leading to the development of models that are unusable in clinical practice. In this work, we evaluate the repeatability of four model types (binary classification, multi-class classification, ordinal classification, and regression) on images that were acquired from the same patient during the same visit. We study the performance of binary, multi-class, ordinal, and regression models on four medical image classification tasks from public and private datasets: knee osteoarthritis, cervical cancer screening, breast density estimation, and retinopathy of prematurity. Repeatability is measured and compared on ResNet and DenseNet architectures. Moreover, we assess the impact of sampling Monte Carlo dropout predictions at test time on classification performance and repeatability. Leveraging Monte Carlo predictions significantly increased repeatability for all tasks on the binary, multi-class, and ordinal models leading to an average reduction of the 95\% limits of agreement by 16% points and of the disagreement rate by 7% points. The classification accuracy improved in most settings along with the repeatability. Our results suggest that beyond about 20 Monte Carlo iterations, there is no further gain in repeatability. In addition to the higher test-retest agreement, Monte Carlo predictions were better calibrated which leads to output probabilities reflecting more accurately the true likelihood of being correctly classified.
Decreasing Annotation Burden of Pairwise Comparisons with Human-in-the-Loop Sorting: Application in Medical Image Artifact Rating
Feb 10, 2022

Abstract:Ranking by pairwise comparisons has shown improved reliability over ordinal classification. However, as the annotations of pairwise comparisons scale quadratically, this becomes less practical when the dataset is large. We propose a method for reducing the number of pairwise comparisons required to rank by a quantitative metric, demonstrating the effectiveness of the approach in ranking medical images by image quality in this proof of concept study. Using the medical image annotation software that we developed, we actively subsample pairwise comparisons using a sorting algorithm with a human rater in the loop. We find that this method substantially reduces the number of comparisons required for a full ordinal ranking without compromising inter-rater reliability when compared to pairwise comparisons without sorting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge