Jaime S. Cardoso
CountPath: Automating Fragment Counting in Digital Pathology
Mar 13, 2025Abstract:Quality control of medical images is a critical component of digital pathology, ensuring that diagnostic images meet required standards. A pre-analytical task within this process is the verification of the number of specimen fragments, a process that ensures that the number of fragments on a slide matches the number documented in the macroscopic report. This step is important to ensure that the slides contain the appropriate diagnostic material from the grossing process, thereby guaranteeing the accuracy of subsequent microscopic examination and diagnosis. Traditionally, this assessment is performed manually, requiring significant time and effort while being subject to significant variability due to its subjective nature. To address these challenges, this study explores an automated approach to fragment counting using the YOLOv9 and Vision Transformer models. Our results demonstrate that the automated system achieves a level of performance comparable to expert assessments, offering a reliable and efficient alternative to manual counting. Additionally, we present findings on interobserver variability, showing that the automated approach achieves an accuracy of 86%, which falls within the range of variation observed among experts (82-88%), further supporting its potential for integration into routine pathology workflows.
An inpainting approach to manipulate asymmetry in pre-operative breast images
Feb 08, 2025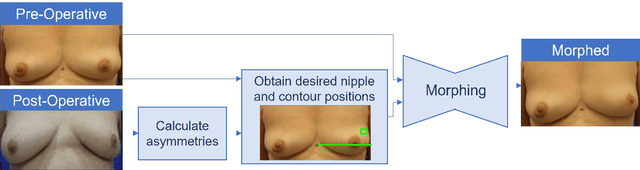
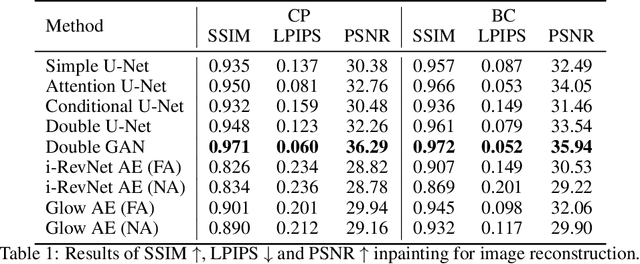
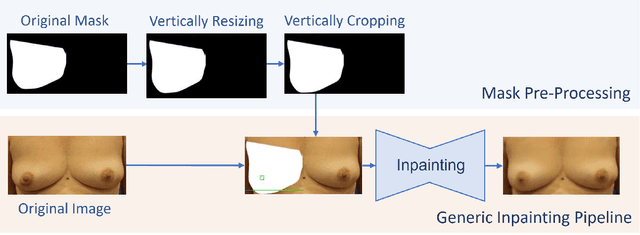

Abstract:One of the most frequent modalities of breast cancer treatment is surgery. Breast surgery can cause visual alterations to the breasts, due to scars and asymmetries. To enable an informed choice of treatment, the patient must be adequately informed of the aesthetic outcomes of each treatment plan. In this work, we propose an inpainting approach to manipulate breast shape and nipple position in breast images, for the purpose of predicting the aesthetic outcomes of breast cancer treatment. We perform experiments with various model architectures for the inpainting task, including invertible networks capable of manipulating breasts in the absence of ground-truth breast contour and nipple annotations. Experiments on two breast datasets show the proposed models' ability to realistically alter a patient's breasts, enabling a faithful reproduction of breast asymmetries of post-operative patients in pre-operative images.
CBVLM: Training-free Explainable Concept-based Large Vision Language Models for Medical Image Classification
Jan 21, 2025



Abstract:The main challenges limiting the adoption of deep learning-based solutions in medical workflows are the availability of annotated data and the lack of interpretability of such systems. Concept Bottleneck Models (CBMs) tackle the latter by constraining the final disease prediction on a set of predefined and human-interpretable concepts. However, the increased interpretability achieved through these concept-based explanations implies a higher annotation burden. Moreover, if a new concept needs to be added, the whole system needs to be retrained. Inspired by the remarkable performance shown by Large Vision-Language Models (LVLMs) in few-shot settings, we propose a simple, yet effective, methodology, CBVLM, which tackles both of the aforementioned challenges. First, for each concept, we prompt the LVLM to answer if the concept is present in the input image. Then, we ask the LVLM to classify the image based on the previous concept predictions. Moreover, in both stages, we incorporate a retrieval module responsible for selecting the best examples for in-context learning. By grounding the final diagnosis on the predicted concepts, we ensure explainability, and by leveraging the few-shot capabilities of LVLMs, we drastically lower the annotation cost. We validate our approach with extensive experiments across four medical datasets and twelve LVLMs (both generic and medical) and show that CBVLM consistently outperforms CBMs and task-specific supervised methods without requiring any training and using just a few annotated examples. More information on our project page: https://cristianopatricio.github.io/CBVLM/.
Second FRCSyn-onGoing: Winning Solutions and Post-Challenge Analysis to Improve Face Recognition with Synthetic Data
Dec 02, 2024Abstract:Synthetic data is gaining increasing popularity for face recognition technologies, mainly due to the privacy concerns and challenges associated with obtaining real data, including diverse scenarios, quality, and demographic groups, among others. It also offers some advantages over real data, such as the large amount of data that can be generated or the ability to customize it to adapt to specific problem-solving needs. To effectively use such data, face recognition models should also be specifically designed to exploit synthetic data to its fullest potential. In order to promote the proposal of novel Generative AI methods and synthetic data, and investigate the application of synthetic data to better train face recognition systems, we introduce the 2nd FRCSyn-onGoing challenge, based on the 2nd Face Recognition Challenge in the Era of Synthetic Data (FRCSyn), originally launched at CVPR 2024. This is an ongoing challenge that provides researchers with an accessible platform to benchmark i) the proposal of novel Generative AI methods and synthetic data, and ii) novel face recognition systems that are specifically proposed to take advantage of synthetic data. We focus on exploring the use of synthetic data both individually and in combination with real data to solve current challenges in face recognition such as demographic bias, domain adaptation, and performance constraints in demanding situations, such as age disparities between training and testing, changes in the pose, or occlusions. Very interesting findings are obtained in this second edition, including a direct comparison with the first one, in which synthetic databases were restricted to DCFace and GANDiffFace.
Classification of Keratitis from Eye Corneal Photographs using Deep Learning
Nov 13, 2024



Abstract:Keratitis is an inflammatory corneal condition responsible for 10% of visual impairment in low- and middle-income countries (LMICs), with bacteria, fungi, or amoeba as the most common infection etiologies. While an accurate and timely diagnosis is crucial for the selected treatment and the patients' sight outcomes, due to the high cost and limited availability of laboratory diagnostics in LMICs, diagnosis is often made by clinical observation alone, despite its lower accuracy. In this study, we investigate and compare different deep learning approaches to diagnose the source of infection: 1) three separate binary models for infection type predictions; 2) a multitask model with a shared backbone and three parallel classification layers (Multitask V1); and, 3) a multitask model with a shared backbone and a multi-head classification layer (Multitask V2). We used a private Brazilian cornea dataset to conduct the empirical evaluation. We achieved the best results with Multitask V2, with an area under the receiver operating characteristic curve (AUROC) confidence intervals of 0.7413-0.7740 (bacteria), 0.8395-0.8725 (fungi), and 0.9448-0.9616 (amoeba). A statistical analysis of the impact of patient features on models' performance revealed that sex significantly affects amoeba infection prediction, and age seems to affect fungi and bacteria predictions.
MST-KD: Multiple Specialized Teachers Knowledge Distillation for Fair Face Recognition
Aug 29, 2024



Abstract:As in school, one teacher to cover all subjects is insufficient to distill equally robust information to a student. Hence, each subject is taught by a highly specialised teacher. Following a similar philosophy, we propose a multiple specialized teacher framework to distill knowledge to a student network. In our approach, directed at face recognition use cases, we train four teachers on one specific ethnicity, leading to four highly specialized and biased teachers. Our strategy learns a project of these four teachers into a common space and distill that information to a student network. Our results highlighted increased performance and reduced bias for all our experiments. In addition, we further show that having biased/specialized teachers is crucial by showing that our approach achieves better results than when knowledge is distilled from four teachers trained on balanced datasets. Our approach represents a step forward to the understanding of the importance of ethnicity-specific features.
Evaluating the Impact of Pulse Oximetry Bias in Machine Learning under Counterfactual Thinking
Aug 08, 2024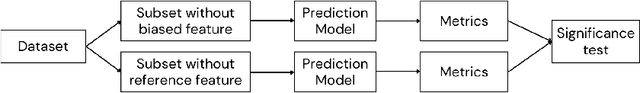
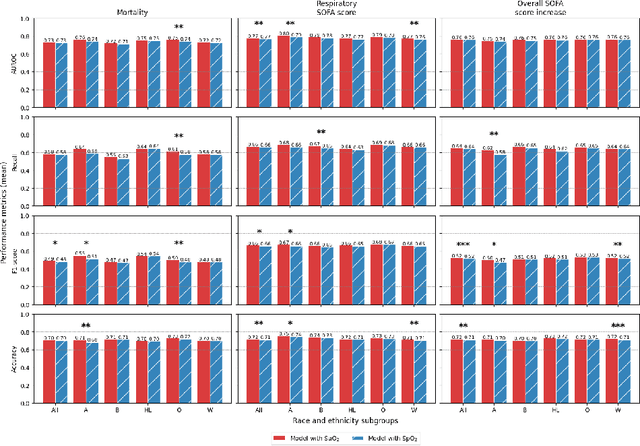

Abstract:Algorithmic bias in healthcare mirrors existing data biases. However, the factors driving unfairness are not always known. Medical devices capture significant amounts of data but are prone to errors; for instance, pulse oximeters overestimate the arterial oxygen saturation of darker-skinned individuals, leading to worse outcomes. The impact of this bias in machine learning (ML) models remains unclear. This study addresses the technical challenges of quantifying the impact of medical device bias in downstream ML. Our experiments compare a "perfect world", without pulse oximetry bias, using SaO2 (blood-gas), to the "actual world", with biased measurements, using SpO2 (pulse oximetry). Under this counterfactual design, two models are trained with identical data, features, and settings, except for the method of measuring oxygen saturation: models using SaO2 are a "control" and models using SpO2 a "treatment". The blood-gas oximetry linked dataset was a suitable test-bed, containing 163,396 nearly-simultaneous SpO2 - SaO2 paired measurements, aligned with a wide array of clinical features and outcomes. We studied three classification tasks: in-hospital mortality, respiratory SOFA score in the next 24 hours, and SOFA score increase by two points. Models using SaO2 instead of SpO2 generally showed better performance. Patients with overestimation of O2 by pulse oximetry of > 3% had significant decreases in mortality prediction recall, from 0.63 to 0.59, P < 0.001. This mirrors clinical processes where biased pulse oximetry readings provide clinicians with false reassurance of patients' oxygen levels. A similar degradation happened in ML models, with pulse oximetry biases leading to more false negatives in predicting adverse outcomes.
Learning Ordinality in Semantic Segmentation
Jul 30, 2024Abstract:Semantic segmentation consists of predicting a semantic label for each image pixel. Conventional deep learning models do not take advantage of ordinal relations that might exist in the domain at hand. For example, it is known that the pupil is inside the iris, and the lane markings are inside the road. Such domain knowledge can be employed as constraints to make the model more robust. The current literature on this topic has explored pixel-wise ordinal segmentation methods, which treat each pixel as an independent observation and promote ordinality in its representation. This paper proposes novel spatial ordinal segmentation methods, which take advantage of the structured image space by considering each pixel as an observation dependent on its neighborhood context to also promote ordinal spatial consistency. When evaluated with five biomedical datasets and multiple configurations of autonomous driving datasets, ordinal methods resulted in more ordinally-consistent models, with substantial improvements in ordinal metrics and some increase in the Dice coefficient. It was also shown that the incorporation of ordinal consistency results in models with better generalization abilities.
A Survey on Cell Nuclei Instance Segmentation and Classification: Leveraging Context and Attention
Jul 26, 2024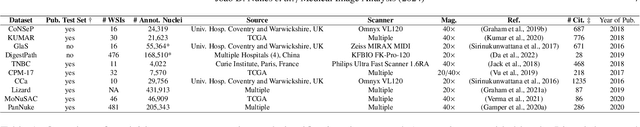
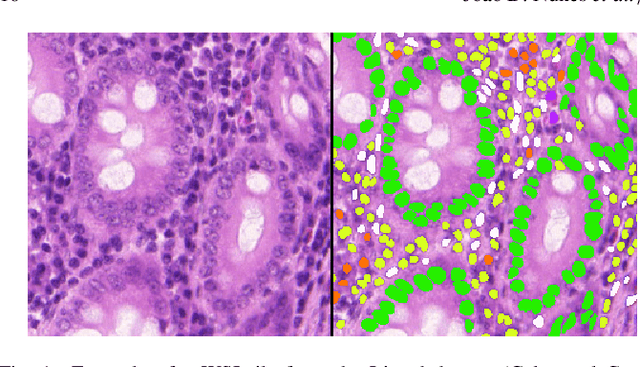
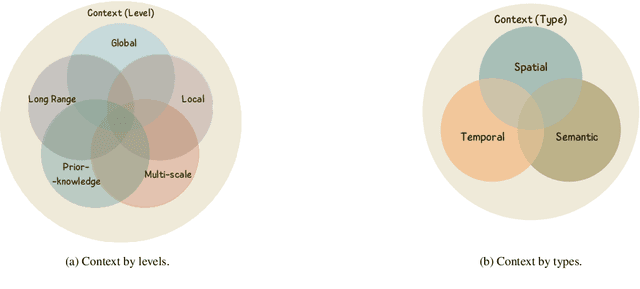
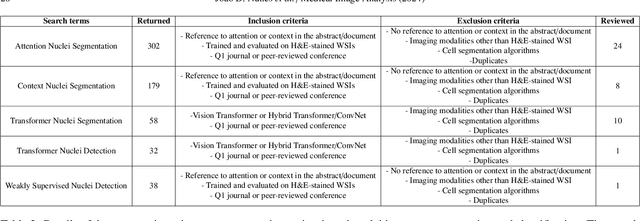
Abstract:Manually annotating nuclei from the gigapixel Hematoxylin and Eosin (H&E)-stained Whole Slide Images (WSIs) is a laborious and costly task, meaning automated algorithms for cell nuclei instance segmentation and classification could alleviate the workload of pathologists and clinical researchers and at the same time facilitate the automatic extraction of clinically interpretable features. But due to high intra- and inter-class variability of nuclei morphological and chromatic features, as well as H&E-stains susceptibility to artefacts, state-of-the-art algorithms cannot correctly detect and classify instances with the necessary performance. In this work, we hypothesise context and attention inductive biases in artificial neural networks (ANNs) could increase the generalization of algorithms for cell nuclei instance segmentation and classification. We conduct a thorough survey on context and attention methods for cell nuclei instance segmentation and classification from H&E-stained microscopy imaging, while providing a comprehensive discussion of the challenges being tackled with context and attention. Besides, we illustrate some limitations of current approaches and present ideas for future research. As a case study, we extend both a general instance segmentation and classification method (Mask-RCNN) and a tailored cell nuclei instance segmentation and classification model (HoVer-Net) with context- and attention-based mechanisms, and do a comparative analysis on a multi-centre colon nuclei identification and counting dataset. Although pathologists rely on context at multiple levels while paying attention to specific Regions of Interest (RoIs) when analysing and annotating WSIs, our findings suggest translating that domain knowledge into algorithm design is no trivial task, but to fully exploit these mechanisms, the scientific understanding of these methods should be addressed.
Deep Learning-based Prediction of Breast Cancer Tumor and Immune Phenotypes from Histopathology
Apr 25, 2024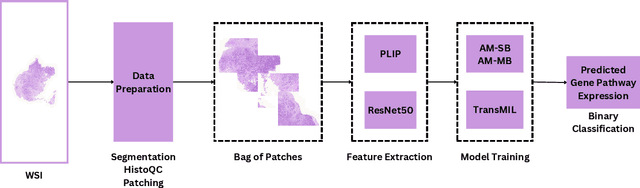

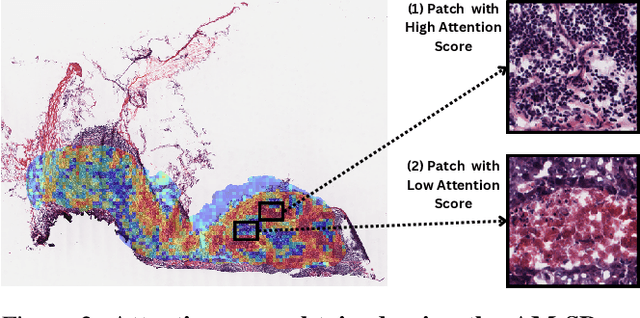

Abstract:The interactions between tumor cells and the tumor microenvironment (TME) dictate therapeutic efficacy of radiation and many systemic therapies in breast cancer. However, to date, there is not a widely available method to reproducibly measure tumor and immune phenotypes for each patient's tumor. Given this unmet clinical need, we applied multiple instance learning (MIL) algorithms to assess activity of ten biologically relevant pathways from the hematoxylin and eosin (H&E) slide of primary breast tumors. We employed different feature extraction approaches and state-of-the-art model architectures. Using binary classification, our models attained area under the receiver operating characteristic (AUROC) scores above 0.70 for nearly all gene expression pathways and on some cases, exceeded 0.80. Attention maps suggest that our trained models recognize biologically relevant spatial patterns of cell sub-populations from H&E. These efforts represent a first step towards developing computational H&E biomarkers that reflect facets of the TME and hold promise for augmenting precision oncology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge