Katharina V. Hoebel
Deep Learning-based Prediction of Breast Cancer Tumor and Immune Phenotypes from Histopathology
Apr 25, 2024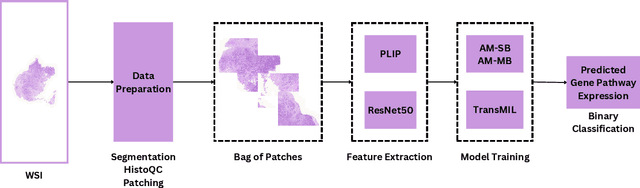

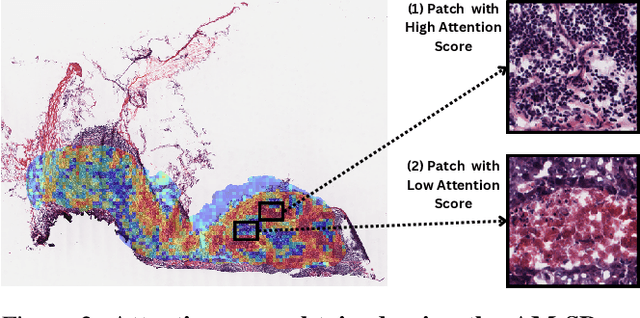

Abstract:The interactions between tumor cells and the tumor microenvironment (TME) dictate therapeutic efficacy of radiation and many systemic therapies in breast cancer. However, to date, there is not a widely available method to reproducibly measure tumor and immune phenotypes for each patient's tumor. Given this unmet clinical need, we applied multiple instance learning (MIL) algorithms to assess activity of ten biologically relevant pathways from the hematoxylin and eosin (H&E) slide of primary breast tumors. We employed different feature extraction approaches and state-of-the-art model architectures. Using binary classification, our models attained area under the receiver operating characteristic (AUROC) scores above 0.70 for nearly all gene expression pathways and on some cases, exceeded 0.80. Attention maps suggest that our trained models recognize biologically relevant spatial patterns of cell sub-populations from H&E. These efforts represent a first step towards developing computational H&E biomarkers that reflect facets of the TME and hold promise for augmenting precision oncology.
A generalized framework to predict continuous scores from medical ordinal labels
May 30, 2023Abstract:Many variables of interest in clinical medicine, like disease severity, are recorded using discrete ordinal categories such as normal/mild/moderate/severe. These labels are used to train and evaluate disease severity prediction models. However, ordinal categories represent a simplification of an underlying continuous severity spectrum. Using continuous scores instead of ordinal categories is more sensitive to detecting small changes in disease severity over time. Here, we present a generalized framework that accurately predicts continuously valued variables using only discrete ordinal labels during model development. We found that for three clinical prediction tasks, models that take the ordinal relationship of the training labels into account outperformed conventional multi-class classification models. Particularly the continuous scores generated by ordinal classification and regression models showed a significantly higher correlation with expert rankings of disease severity and lower mean squared errors compared to the multi-class classification models. Furthermore, the use of MC dropout significantly improved the ability of all evaluated deep learning approaches to predict continuously valued scores that truthfully reflect the underlying continuous target variable. We showed that accurate continuously valued predictions can be generated even if the model development only involves discrete ordinal labels. The novel framework has been validated on three different clinical prediction tasks and has proven to bridge the gap between discrete ordinal labels and the underlying continuously valued variables.
Federated Learning for Breast Density Classification: A Real-World Implementation
Sep 17, 2020

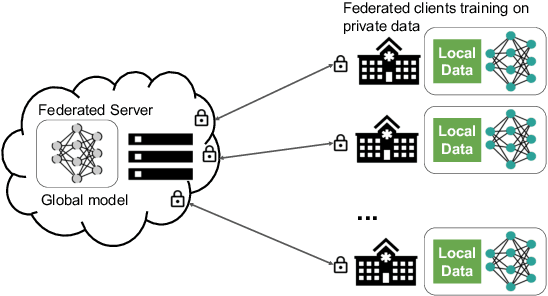
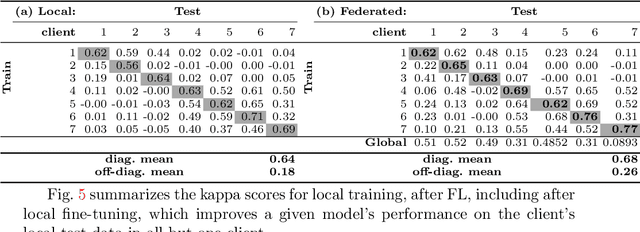
Abstract:Building robust deep learning-based models requires large quantities of diverse training data. In this study, we investigate the use of federated learning (FL) to build medical imaging classification models in a real-world collaborative setting. Seven clinical institutions from across the world joined this FL effort to train a model for breast density classification based on Breast Imaging, Reporting & Data System (BI-RADS). We show that despite substantial differences among the datasets from all sites (mammography system, class distribution, and data set size) and without centralizing data, we can successfully train AI models in federation. The results show that models trained using FL perform 6.3% on average better than their counterparts trained on an institute's local data alone. Furthermore, we show a 45.8% relative improvement in the models' generalizability when evaluated on the other participating sites' testing data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge