Susan Ostmo
A generalized framework to predict continuous scores from medical ordinal labels
May 30, 2023Abstract:Many variables of interest in clinical medicine, like disease severity, are recorded using discrete ordinal categories such as normal/mild/moderate/severe. These labels are used to train and evaluate disease severity prediction models. However, ordinal categories represent a simplification of an underlying continuous severity spectrum. Using continuous scores instead of ordinal categories is more sensitive to detecting small changes in disease severity over time. Here, we present a generalized framework that accurately predicts continuously valued variables using only discrete ordinal labels during model development. We found that for three clinical prediction tasks, models that take the ordinal relationship of the training labels into account outperformed conventional multi-class classification models. Particularly the continuous scores generated by ordinal classification and regression models showed a significantly higher correlation with expert rankings of disease severity and lower mean squared errors compared to the multi-class classification models. Furthermore, the use of MC dropout significantly improved the ability of all evaluated deep learning approaches to predict continuously valued scores that truthfully reflect the underlying continuous target variable. We showed that accurate continuously valued predictions can be generated even if the model development only involves discrete ordinal labels. The novel framework has been validated on three different clinical prediction tasks and has proven to bridge the gap between discrete ordinal labels and the underlying continuously valued variables.
Not Color Blind: AI Predicts Racial Identity from Black and White Retinal Vessel Segmentations
Sep 28, 2021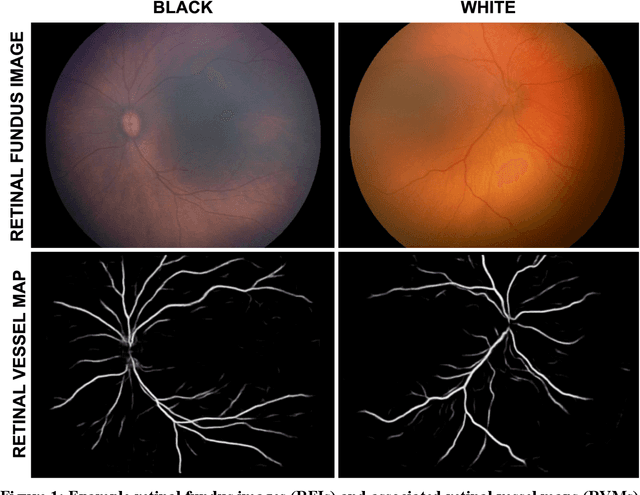
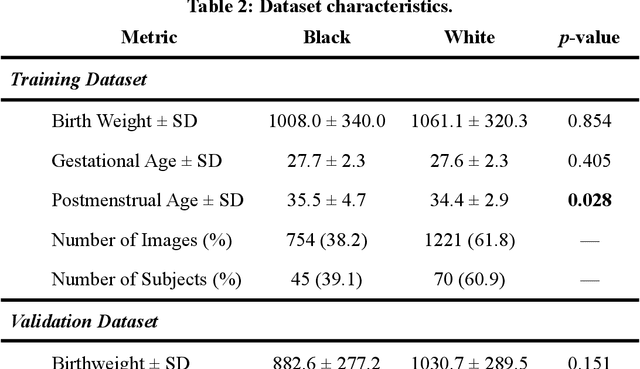
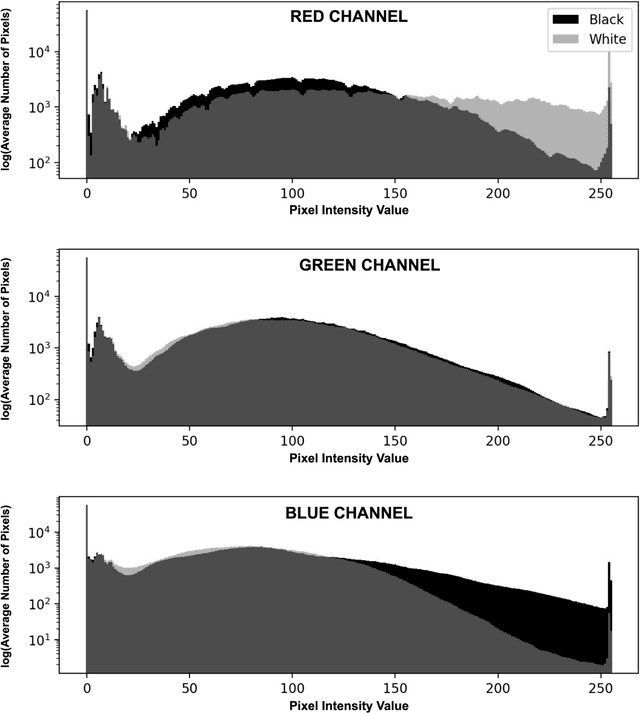
Abstract:Background: Artificial intelligence (AI) may demonstrate racial bias when skin or choroidal pigmentation is present in medical images. Recent studies have shown that convolutional neural networks (CNNs) can predict race from images that were not previously thought to contain race-specific features. We evaluate whether grayscale retinal vessel maps (RVMs) of patients screened for retinopathy of prematurity (ROP) contain race-specific features. Methods: 4095 retinal fundus images (RFIs) were collected from 245 Black and White infants. A U-Net generated RVMs from RFIs, which were subsequently thresholded, binarized, or skeletonized. To determine whether RVM differences between Black and White eyes were physiological, CNNs were trained to predict race from color RFIs, raw RVMs, and thresholded, binarized, or skeletonized RVMs. Area under the precision-recall curve (AUC-PR) was evaluated. Findings: CNNs predicted race from RFIs near perfectly (image-level AUC-PR: 0.999, subject-level AUC-PR: 1.000). Raw RVMs were almost as informative as color RFIs (image-level AUC-PR: 0.938, subject-level AUC-PR: 0.995). Ultimately, CNNs were able to detect whether RFIs or RVMs were from Black or White babies, regardless of whether images contained color, vessel segmentation brightness differences were nullified, or vessel segmentation widths were normalized. Interpretation: AI can detect race from grayscale RVMs that were not thought to contain racial information. Two potential explanations for these findings are that: retinal vessels physiologically differ between Black and White babies or the U-Net segments the retinal vasculature differently for various fundus pigmentations. Either way, the implications remain the same: AI algorithms have potential to demonstrate racial bias in practice, even when preliminary attempts to remove such information from the underlying images appear to be successful.
Accelerated Experimental Design for Pairwise Comparisons
Jan 18, 2019
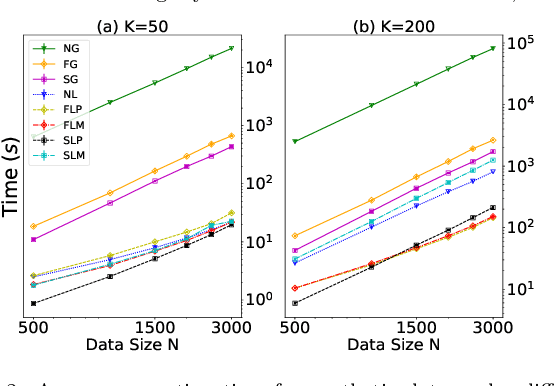
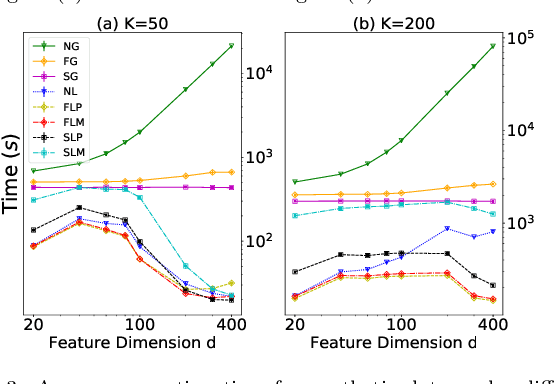
Abstract:Pairwise comparison labels are more informative and less variable than class labels, but generating them poses a challenge: their number grows quadratically in the dataset size. We study a natural experimental design objective, namely, D-optimality, that can be used to identify which $K$ pairwise comparisons to generate. This objective is known to perform well in practice, and is submodular, making the selection approximable via the greedy algorithm. A na\"ive greedy implementation has $O(N^2d^2K)$ complexity, where $N$ is the dataset size, $d$ is the feature space dimension, and $K$ is the number of generated comparisons. We show that, by exploiting the inherent geometry of the dataset--namely, that it consists of pairwise comparisons--the greedy algorithm's complexity can be reduced to $O(N^2(K+d)+N(dK+d^2) +d^2K).$ We apply the same acceleration also to the so-called lazy greedy algorithm. When combined, the above improvements lead to an execution time of less than 1 hour for a dataset with $10^8$ comparisons; the na\"ive greedy algorithm on the same dataset would require more than 10 days to terminate.
High-resolution medical image synthesis using progressively grown generative adversarial networks
May 09, 2018
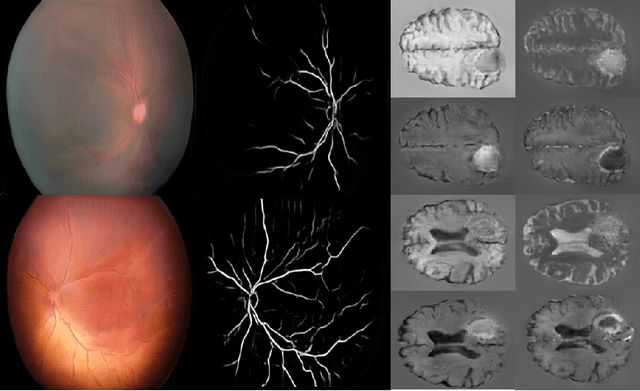
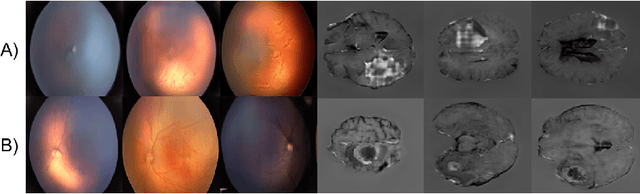
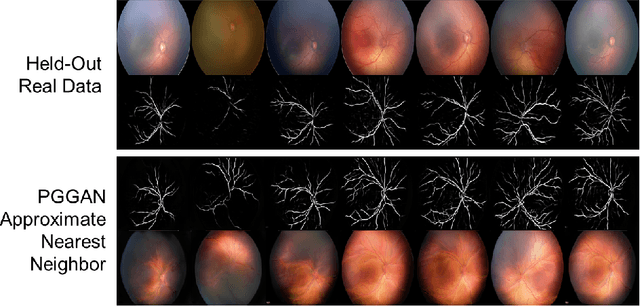
Abstract:Generative adversarial networks (GANs) are a class of unsupervised machine learning algorithms that can produce realistic images from randomly-sampled vectors in a multi-dimensional space. Until recently, it was not possible to generate realistic high-resolution images using GANs, which has limited their applicability to medical images that contain biomarkers only detectable at native resolution. Progressive growing of GANs is an approach wherein an image generator is trained to initially synthesize low resolution synthetic images (8x8 pixels), which are then fed to a discriminator that distinguishes these synthetic images from real downsampled images. Additional convolutional layers are then iteratively introduced to produce images at twice the previous resolution until the desired resolution is reached. In this work, we demonstrate that this approach can produce realistic medical images in two different domains; fundus photographs exhibiting vascular pathology associated with retinopathy of prematurity (ROP), and multi-modal magnetic resonance images of glioma. We also show that fine-grained details associated with pathology, such as retinal vessels or tumor heterogeneity, can be preserved and enhanced by including segmentation maps as additional channels. We envisage several applications of the approach, including image augmentation and unsupervised classification of pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge