James Brown
RoLID-11K: A Dashcam Dataset for Small-Object Roadside Litter Detection
Jan 01, 2026Abstract:Roadside litter poses environmental, safety and economic challenges, yet current monitoring relies on labour-intensive surveys and public reporting, providing limited spatial coverage. Existing vision datasets for litter detection focus on street-level still images, aerial scenes or aquatic environments, and do not reflect the unique characteristics of dashcam footage, where litter appears extremely small, sparse and embedded in cluttered road-verge backgrounds. We introduce RoLID-11K, the first large-scale dataset for roadside litter detection from dashcams, comprising over 11k annotated images spanning diverse UK driving conditions and exhibiting pronounced long-tail and small-object distributions. We benchmark a broad spectrum of modern detectors, from accuracy-oriented transformer architectures to real-time YOLO models, and analyse their strengths and limitations on this challenging task. Our results show that while CO-DETR and related transformers achieve the best localisation accuracy, real-time models remain constrained by coarse feature hierarchies. RoLID-11K establishes a challenging benchmark for extreme small-object detection in dynamic driving scenes and aims to support the development of scalable, low-cost systems for roadside-litter monitoring. The dataset is available at https://github.com/xq141839/RoLID-11K.
MMDEW: Multipurpose Multiclass Density Estimation in the Wild
Oct 02, 2025Abstract:Density map estimation can be used to estimate object counts in dense and occluded scenes where discrete counting-by-detection methods fail. We propose a multicategory counting framework that leverages a Twins pyramid vision-transformer backbone and a specialised multi-class counting head built on a state-of-the-art multiscale decoding approach. A two-task design adds a segmentation-based Category Focus Module, suppressing inter-category cross-talk at training time. Training and evaluation on the VisDrone and iSAID benchmarks demonstrates superior performance versus prior multicategory crowd-counting approaches (33%, 43% and 64% reduction to MAE), and the comparison with YOLOv11 underscores the necessity of crowd counting methods in dense scenes. The method's regional loss opens up multi-class crowd counting to new domains, demonstrated through the application to a biodiversity monitoring dataset, highlighting its capacity to inform conservation efforts and enable scalable ecological insights.
Unsupervised Tomato Split Anomaly Detection using Hyperspectral Imaging and Variational Autoencoders
Jan 06, 2025



Abstract:Tomato anomalies/damages pose a significant challenge in greenhouse farming. While this method of cultivation benefits from efficient resource utilization, anomalies can significantly degrade the quality of farm produce. A common anomaly associated with tomatoes is splitting, characterized by the development of cracks on the tomato skin, which degrades its quality. Detecting this type of anomaly is challenging due to dynamic variations in appearance and sizes, compounded by dataset scarcity. We address this problem in an unsupervised manner by utilizing a tailored variational autoencoder (VAE) with hyperspectral input. Preliminary analysis of the dataset enabled us to select the optimal range of wavelengths for detecting this anomaly. Our findings indicate that the 530nm - 550nm range is suitable for identifying tomato dry splits. The analysis on reconstruction loss allow us to not only detect the anomalies but also to some degree estimate the anomalous regions.
FReSCO: Flow Reconstruction and Segmentation for low latency Cardiac Output monitoring using deep artifact suppression and segmentation
Mar 25, 2022



Abstract:Purpose: Real-time monitoring of cardiac output (CO) requires low latency reconstruction and segmentation of real-time phase contrast MR (PCMR), which has previously been difficult to perform. Here we propose a deep learning framework for 'Flow Reconstruction and Segmentation for low latency Cardiac Output monitoring' (FReSCO). Methods: Deep artifact suppression and segmentation U-Nets were independently trained. Breath hold spiral PCMR data (n=516) was synthetically undersampled using a variable density spiral sampling pattern and gridded to create aliased data for training of the artifact suppression U-net. A subset of the data (n=96) was segmented and used to train the segmentation U-net. Real-time spiral PCMR was prospectively acquired and then reconstructed and segmented using the trained models (FReSCO) at low latency at the scanner in 10 healthy subjects during rest, exercise and recovery periods. CO obtained via FReSCO was compared to a reference rest CO and rest and exercise Compressed Sensing (CS) CO. Results: FReSCO was demonstrated prospectively at the scanner. Beat-to-beat heartrate, stroke volume and CO could be visualized with a mean latency of 622ms. No significant differences were noted when compared to reference at rest (Bias = -0.21+-0.50 L/min, p=0.246) or CS at peak exercise (Bias=0.12+-0.48 L/min, p=0.458). Conclusion: FReSCO was successfully demonstrated for real-time monitoring of CO during exercise and could provide a convenient tool for assessment of the hemodynamic response to a range of stressors.
DeepNeuro: an open-source deep learning toolbox for neuroimaging
Aug 14, 2018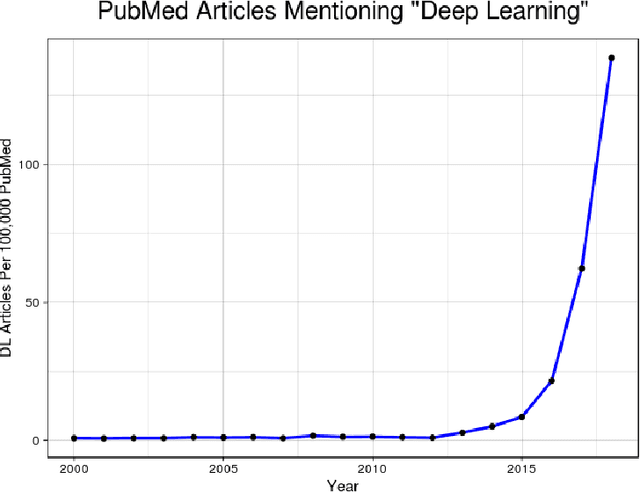
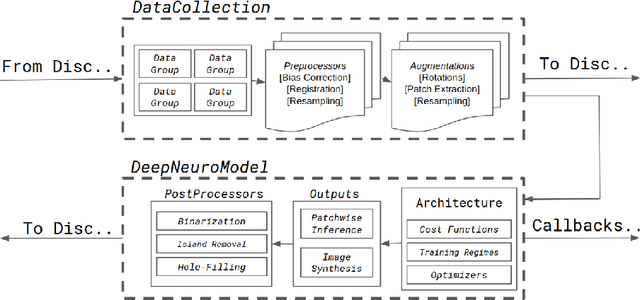
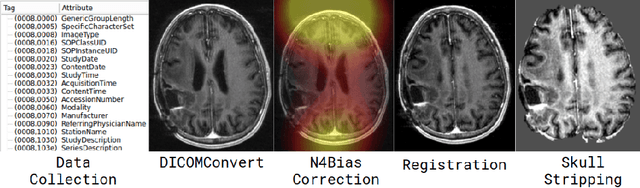
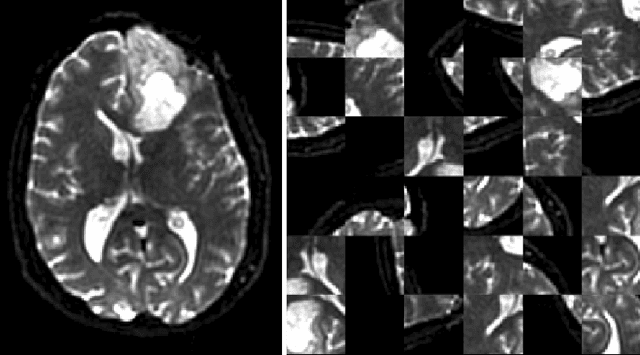
Abstract:Translating neural networks from theory to clinical practice has unique challenges, specifically in the field of neuroimaging. In this paper, we present DeepNeuro, a deep learning framework that is best-suited to putting deep learning algorithms for neuroimaging in practical usage with a minimum of friction. We show how this framework can be used to both design and train neural network architectures, as well as modify state-of-the-art architectures in a flexible and intuitive way. We display the pre- and postprocessing functions common in the medical imaging community that DeepNeuro offers to ensure consistent performance of networks across variable users, institutions, and scanners. And we show how pipelines created in DeepNeuro can be concisely packaged into shareable Docker containers and command-line interfaces using DeepNeuro's pipeline resources.
High-resolution medical image synthesis using progressively grown generative adversarial networks
May 09, 2018
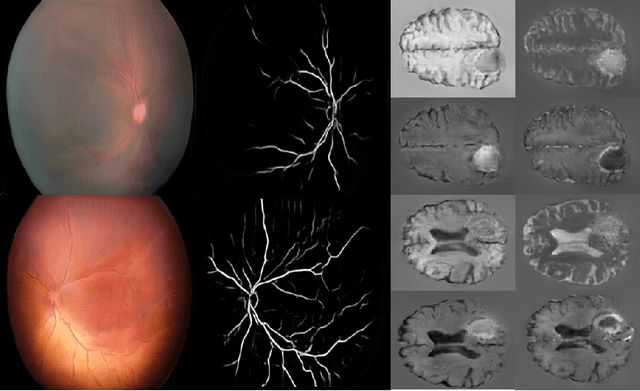
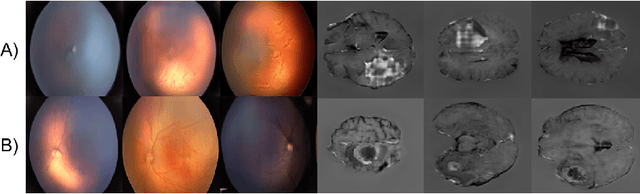
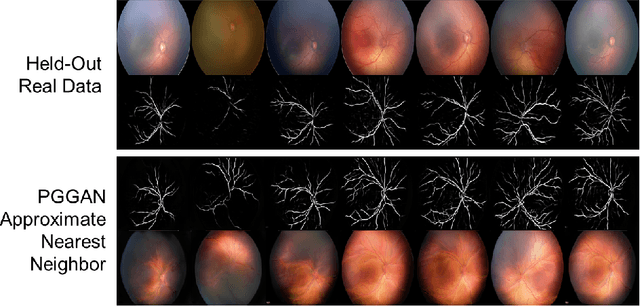
Abstract:Generative adversarial networks (GANs) are a class of unsupervised machine learning algorithms that can produce realistic images from randomly-sampled vectors in a multi-dimensional space. Until recently, it was not possible to generate realistic high-resolution images using GANs, which has limited their applicability to medical images that contain biomarkers only detectable at native resolution. Progressive growing of GANs is an approach wherein an image generator is trained to initially synthesize low resolution synthetic images (8x8 pixels), which are then fed to a discriminator that distinguishes these synthetic images from real downsampled images. Additional convolutional layers are then iteratively introduced to produce images at twice the previous resolution until the desired resolution is reached. In this work, we demonstrate that this approach can produce realistic medical images in two different domains; fundus photographs exhibiting vascular pathology associated with retinopathy of prematurity (ROP), and multi-modal magnetic resonance images of glioma. We also show that fine-grained details associated with pathology, such as retinal vessels or tumor heterogeneity, can be preserved and enhanced by including segmentation maps as additional channels. We envisage several applications of the approach, including image augmentation and unsupervised classification of pathology.
Sequential 3D U-Nets for Biologically-Informed Brain Tumor Segmentation
Sep 09, 2017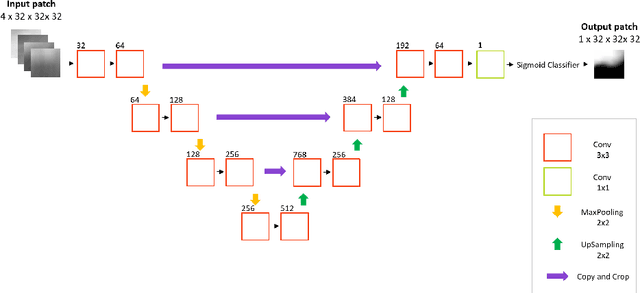

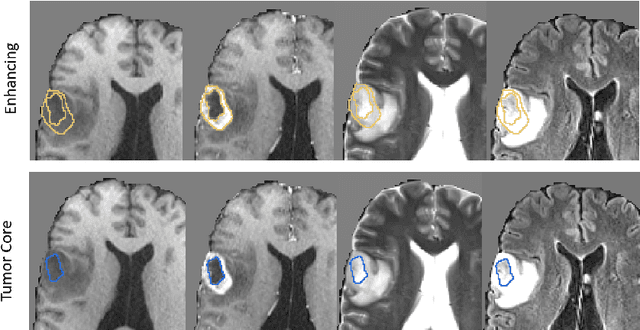
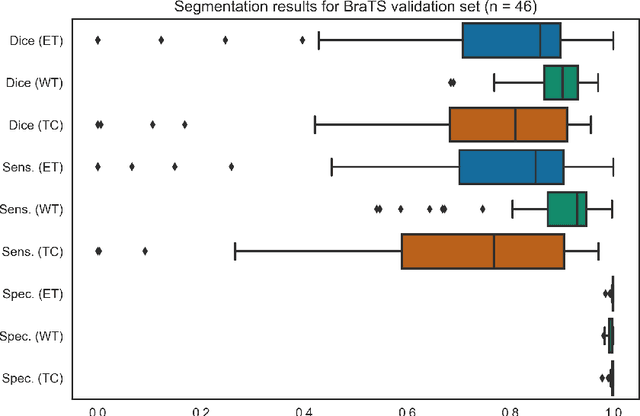
Abstract:Deep learning has quickly become the weapon of choice for brain lesion segmentation. However, few existing algorithms pre-configure any biological context of their chosen segmentation tissues, and instead rely on the neural network's optimizer to develop such associations de novo. We present a novel method for applying deep neural networks to the problem of glioma tissue segmentation that takes into account the structured nature of gliomas - edematous tissue surrounding mutually-exclusive regions of enhancing and non-enhancing tumor. We trained multiple deep neural networks with a 3D U-Net architecture in a tree structure to create segmentations for edema, non-enhancing tumor, and enhancing tumor regions. Specifically, training was configured such that the whole tumor region including edema was predicted first, and its output segmentation was fed as input into separate models to predict enhancing and non-enhancing tumor. Our method was trained and evaluated on the publicly available BraTS dataset, achieving Dice scores of 0.882, 0.732, and 0.730 for whole tumor, enhancing tumor and tumor core respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge