Simon Arridge
Learning Binary Sampling Patterns for Single-Pixel Imaging using Bilevel Optimisation
Aug 26, 2025Abstract:Single-Pixel Imaging enables reconstructing objects using a single detector through sequential illuminations with structured light patterns. We propose a bilevel optimisation method for learning task-specific, binary illumination patterns, optimised for applications like single-pixel fluorescence microscopy. We address the non-differentiable nature of binary pattern optimisation using the Straight-Through Estimator and leveraging a Total Deep Variation regulariser in the bilevel formulation. We demonstrate our method on the CytoImageNet microscopy dataset and show that learned patterns achieve superior reconstruction performance compared to baseline methods, especially in highly undersampled regimes.
Plug-and-Play Half-Quadratic Splitting for Ptychography
Dec 03, 2024



Abstract:Ptychography is a coherent diffraction imaging method that uses phase retrieval techniques to reconstruct complex-valued images. It achieves this by sequentially illuminating overlapping regions of a sample with a coherent beam and recording the diffraction pattern. Although this addresses traditional imaging system challenges, it is computationally intensive and highly sensitive to noise, especially with reduced illumination overlap. Data-driven regularisation techniques have been applied in phase retrieval to improve reconstruction quality. In particular, plug-and-play (PnP) offers flexibility by integrating data-driven denoisers as implicit priors. In this work, we propose a half-quadratic splitting framework for using PnP and other data-driven priors for ptychography. We evaluate our method both on natural images and real test objects to validate its effectiveness for ptychographic image reconstruction.
Equidistribution-based training of Free Knot Splines and ReLU Neural Networks
Jul 02, 2024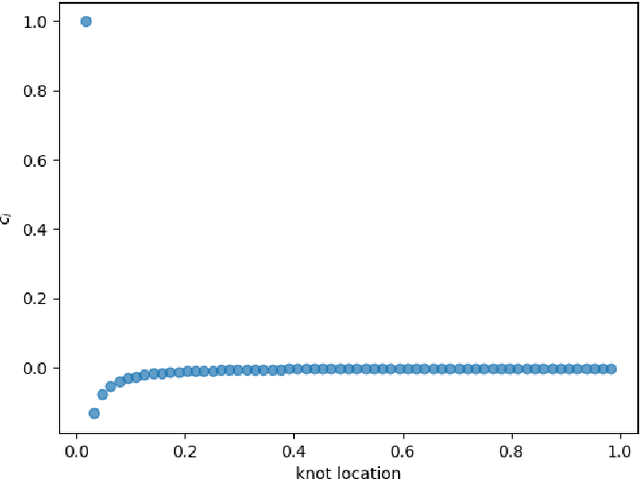

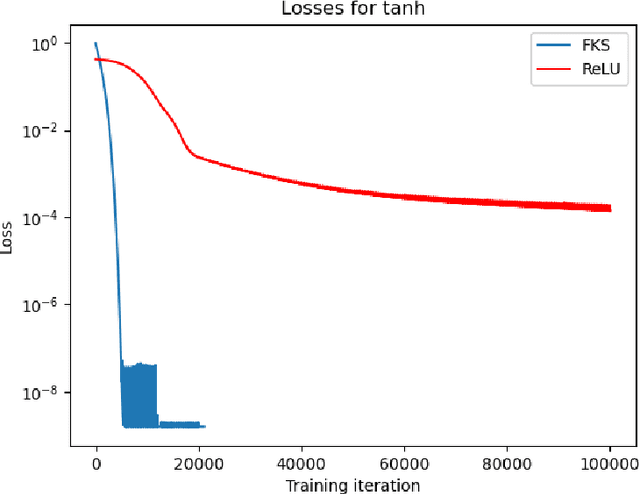
Abstract:We consider the problem of one-dimensional function approximation using shallow neural networks (NN) with a rectified linear unit (ReLU) activation function and compare their training with traditional methods such as univariate Free Knot Splines (FKS). ReLU NNs and FKS span the same function space, and thus have the same theoretical expressivity. In the case of ReLU NNs, we show that their ill-conditioning degrades rapidly as the width of the network increases. This often leads to significantly poorer approximation in contrast to the FKS representation, which remains well-conditioned as the number of knots increases. We leverage the theory of optimal piecewise linear interpolants to improve the training procedure for a ReLU NN. Using the equidistribution principle, we propose a two-level procedure for training the FKS by first solving the nonlinear problem of finding the optimal knot locations of the interpolating FKS. Determining the optimal knots then acts as a good starting point for training the weights of the FKS. The training of the FKS gives insights into how we can train a ReLU NN effectively to give an equally accurate approximation. More precisely, we combine the training of the ReLU NN with an equidistribution based loss to find the breakpoints of the ReLU functions, combined with preconditioning the ReLU NN approximation (to take an FKS form) to find the scalings of the ReLU functions, leads to a well-conditioned and reliable method of finding an accurate ReLU NN approximation to a target function. We test this method on a series or regular, singular, and rapidly varying target functions and obtain good results realising the expressivity of the network in this case.
Learning Governing Equations of Unobserved States in Dynamical Systems
Apr 29, 2024Abstract:Data driven modelling and scientific machine learning have been responsible for significant advances in determining suitable models to describe data. Within dynamical systems, neural ordinary differential equations (ODEs), where the system equations are set to be governed by a neural network, have become a popular tool for this challenge in recent years. However, less emphasis has been placed on systems that are only partially-observed. In this work, we employ a hybrid neural ODE structure, where the system equations are governed by a combination of a neural network and domain-specific knowledge, together with symbolic regression (SR), to learn governing equations of partially-observed dynamical systems. We test this approach on two case studies: A 3-dimensional model of the Lotka-Volterra system and a 5-dimensional model of the Lorenz system. We demonstrate that the method is capable of successfully learning the true underlying governing equations of unobserved states within these systems, with robustness to measurement noise.
Investigating the use of publicly available natural videos to learn Dynamic MR image reconstruction
Nov 23, 2023Abstract:Purpose: To develop and assess a deep learning (DL) pipeline to learn dynamic MR image reconstruction from publicly available natural videos (Inter4K). Materials and Methods: Learning was performed for a range of DL architectures (VarNet, 3D UNet, FastDVDNet) and corresponding sampling patterns (Cartesian, radial, spiral) either from true multi-coil cardiac MR data (N=692) or from pseudo-MR data simulated from Inter4K natural videos (N=692). Real-time undersampled dynamic MR images were reconstructed using DL networks trained with cardiac data and natural videos, and compressed sensing (CS). Differences were assessed in simulations (N=104 datasets) in terms of MSE, PSNR, and SSIM and prospectively for cardiac (short axis, four chambers, N=20) and speech (N=10) data in terms of subjective image quality ranking, SNR and Edge sharpness. Friedman Chi Square tests with post-hoc Nemenyi analysis were performed to assess statistical significance. Results: For all simulation metrics, DL networks trained with cardiac data outperformed DL networks trained with natural videos, which outperformed CS (p<0.05). However, in prospective experiments DL reconstructions using both training datasets were ranked similarly (and higher than CS) and presented no statistical differences in SNR and Edge Sharpness for most conditions. Additionally, high SSIM was measured between the DL methods with cardiac data and natural videos (SSIM>0.85). Conclusion: The developed pipeline enabled learning dynamic MR reconstruction from natural videos preserving DL reconstruction advantages such as high quality fast and ultra-fast reconstructions while overcoming some limitations (data scarcity or sharing). The natural video dataset, code and pre-trained networks are made readily available on github. Key Words: real-time; dynamic MRI; deep learning; image reconstruction; machine learning;
Inverse Problems with Learned Forward Operators
Nov 21, 2023



Abstract:Solving inverse problems requires knowledge of the forward operator, but accurate models can be computationally expensive and hence cheaper variants are desired that do not compromise reconstruction quality. This chapter reviews reconstruction methods in inverse problems with learned forward operators that follow two different paradigms. The first one is completely agnostic to the forward operator and learns its restriction to the subspace spanned by the training data. The framework of regularisation by projection is then used to find a reconstruction. The second one uses a simplified model of the physics of the measurement process and only relies on the training data to learn a model correction. We present the theory of these two approaches and compare them numerically. A common theme emerges: both methods require, or at least benefit from, training data not only for the forward operator, but also for its adjoint.
Score-Based Generative Models for PET Image Reconstruction
Aug 27, 2023



Abstract:Score-based generative models have demonstrated highly promising results for medical image reconstruction tasks in magnetic resonance imaging or computed tomography. However, their application to Positron Emission Tomography (PET) is still largely unexplored. PET image reconstruction involves a variety of challenges, including Poisson noise with high variance and a wide dynamic range. To address these challenges, we propose several PET-specific adaptations of score-based generative models. The proposed framework is developed for both 2D and 3D PET. In addition, we provide an extension to guided reconstruction using magnetic resonance images. We validate the approach through extensive 2D and 3D $\textit{in-silico}$ experiments with a model trained on patient-realistic data without lesions, and evaluate on data without lesions as well as out-of-distribution data with lesions. This demonstrates the proposed method's robustness and significant potential for improved PET reconstruction.
A Learned Born Series for Highly-Scattering Media
Dec 09, 2022
Abstract:A new method for solving the wave equation is presented, called the learned Born series (LBS), which is derived from a convergent Born Series but its components are found through training. The LBS is shown to be significantly more accurate than the convergent Born series for the same number of iterations, in the presence of high contrast scatterers, while maintaining a comparable computational complexity. The LBS is able to generate a reasonable prediction of the global pressure field with a small number of iterations, and the errors decrease with the number of learned iterations.
Unsupervised denoising for sparse multi-spectral computed tomography
Nov 02, 2022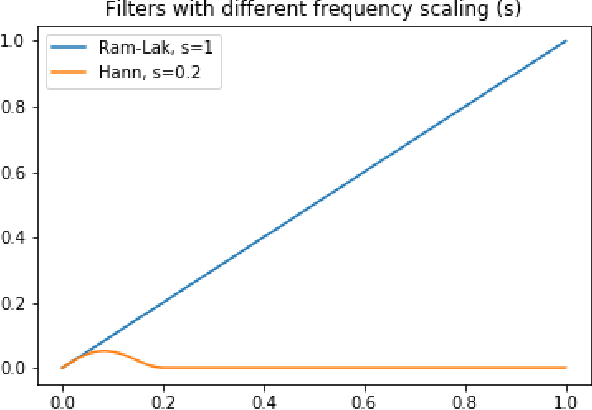
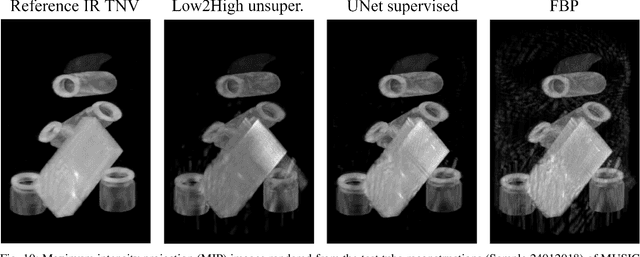
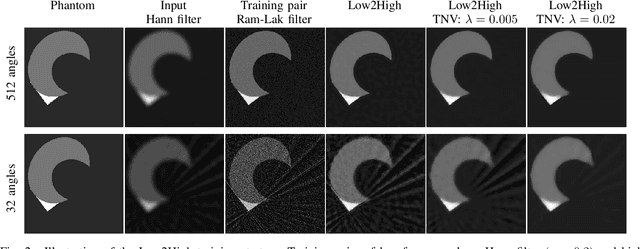
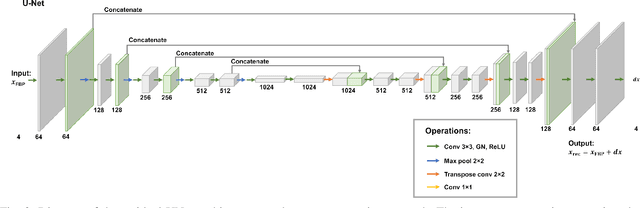
Abstract:Multi-energy computed tomography (CT) with photon counting detectors (PCDs) enables spectral imaging as PCDs can assign the incoming photons to specific energy channels. However, PCDs with many spectral channels drastically increase the computational complexity of the CT reconstruction, and bespoke reconstruction algorithms need fine-tuning to varying noise statistics. \rev{Especially if many projections are taken, a large amount of data has to be collected and stored. Sparse view CT is one solution for data reduction. However, these issues are especially exacerbated when sparse imaging scenarios are encountered due to a significant reduction in photon counts.} In this work, we investigate the suitability of learning-based improvements to the challenging task of obtaining high-quality reconstructions from sparse measurements for a 64-channel PCD-CT. In particular, to overcome missing reference data for the training procedure, we propose an unsupervised denoising and artefact removal approach by exploiting different filter functions in the reconstruction and an explicit coupling of spectral channels with the nuclear norm. Performance is assessed on both simulated synthetic data and the openly available experimental Multi-Spectral Imaging via Computed Tomography (MUSIC) dataset. We compared the quality of our unsupervised method to iterative total nuclear variation regularized reconstructions and a supervised denoiser trained with reference data. We show that improved reconstruction quality can be achieved with flexibility on noise statistics and effective suppression of streaking artefacts when using unsupervised denoising with spectral coupling.
FReSCO: Flow Reconstruction and Segmentation for low latency Cardiac Output monitoring using deep artifact suppression and segmentation
Mar 25, 2022



Abstract:Purpose: Real-time monitoring of cardiac output (CO) requires low latency reconstruction and segmentation of real-time phase contrast MR (PCMR), which has previously been difficult to perform. Here we propose a deep learning framework for 'Flow Reconstruction and Segmentation for low latency Cardiac Output monitoring' (FReSCO). Methods: Deep artifact suppression and segmentation U-Nets were independently trained. Breath hold spiral PCMR data (n=516) was synthetically undersampled using a variable density spiral sampling pattern and gridded to create aliased data for training of the artifact suppression U-net. A subset of the data (n=96) was segmented and used to train the segmentation U-net. Real-time spiral PCMR was prospectively acquired and then reconstructed and segmented using the trained models (FReSCO) at low latency at the scanner in 10 healthy subjects during rest, exercise and recovery periods. CO obtained via FReSCO was compared to a reference rest CO and rest and exercise Compressed Sensing (CS) CO. Results: FReSCO was demonstrated prospectively at the scanner. Beat-to-beat heartrate, stroke volume and CO could be visualized with a mean latency of 622ms. No significant differences were noted when compared to reference at rest (Bias = -0.21+-0.50 L/min, p=0.246) or CS at peak exercise (Bias=0.12+-0.48 L/min, p=0.458). Conclusion: FReSCO was successfully demonstrated for real-time monitoring of CO during exercise and could provide a convenient tool for assessment of the hemodynamic response to a range of stressors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge