Peter Maass
University of Bremen, aisencia
Deep Learning Based Reconstruction Methods for Electrical Impedance Tomography
Aug 08, 2025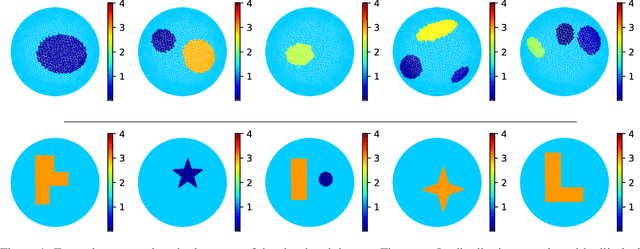
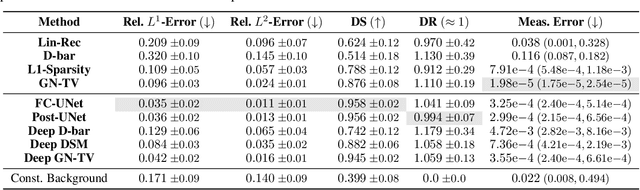
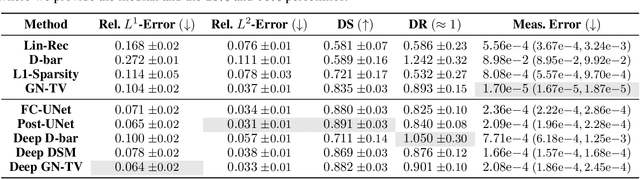
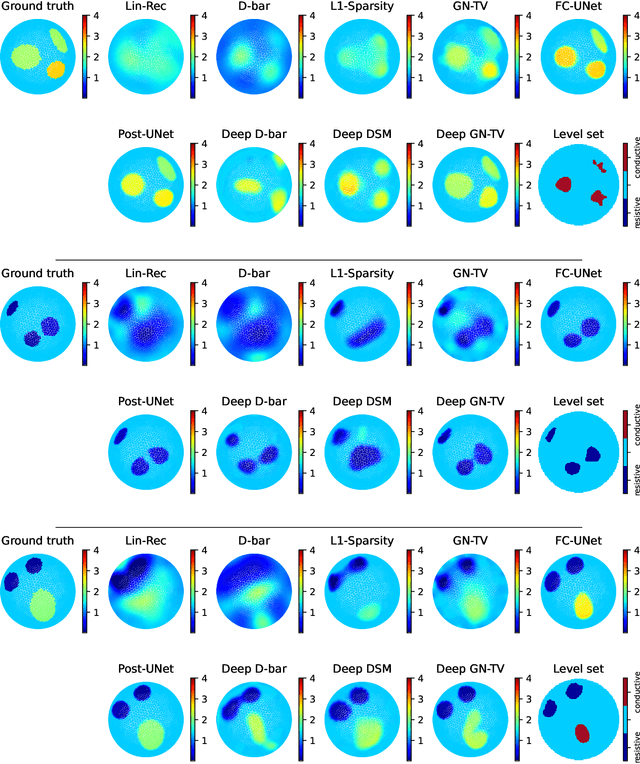
Abstract:Electrical Impedance Tomography (EIT) is a powerful imaging modality widely used in medical diagnostics, industrial monitoring, and environmental studies. The EIT inverse problem is about inferring the internal conductivity distribution of the concerned object from the voltage measurements taken on its boundary. This problem is severely ill-posed, and requires advanced computational approaches for accurate and reliable image reconstruction. Recent innovations in both model-based reconstruction and deep learning have driven significant progress in the field. In this review, we explore learned reconstruction methods that employ deep neural networks for solving the EIT inverse problem. The discussion focuses on the complete electrode model, one popular mathematical model for real-world applications of EIT. We compare a wide variety of learned approaches, including fully-learned, post-processing and learned iterative methods, with several conventional model-based reconstruction techniques, e.g., sparsity regularization, regularized Gauss-Newton iteration and level set method. The evaluation is based on three datasets: a simulated dataset of ellipses, an out-of-distribution simulated dataset, and the KIT4 dataset, including real-world measurements. Our results demonstrate that learned methods outperform model-based methods for in-distribution data but face challenges in generalization, where hybrid methods exhibit a good balance of accuracy and adaptability.
Visualize and Paint GAN Activations
May 24, 2024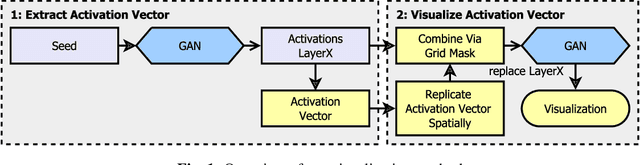
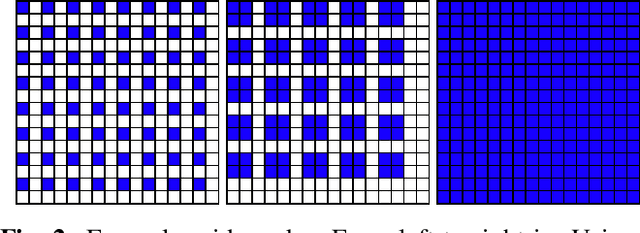
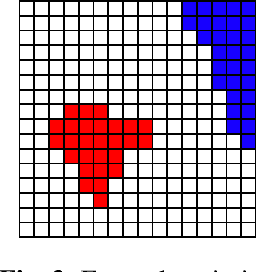
Abstract:We investigate how generated structures of GANs correlate with their activations in hidden layers, with the purpose of better understanding the inner workings of those models and being able to paint structures with unconditionally trained GANs. This gives us more control over the generated images, allowing to generate them from a semantic segmentation map while not requiring such a segmentation in the training data. To this end we introduce the concept of tileable features, allowing us to identify activations that work well for painting.
Enhancing the analysis of murine neonatal ultrasonic vocalizations: Development, evaluation, and application of different mathematical models
May 17, 2024Abstract:Rodents employ a broad spectrum of ultrasonic vocalizations (USVs) for social communication. As these vocalizations offer valuable insights into affective states, social interactions, and developmental stages of animals, various deep learning approaches have aimed to automate both the quantitative (detection) and qualitative (classification) analysis of USVs. Here, we present the first systematic evaluation of different types of neural networks for USV classification. We assessed various feedforward networks, including a custom-built, fully-connected network and convolutional neural network, different residual neural networks (ResNets), an EfficientNet, and a Vision Transformer (ViT). Paired with a refined, entropy-based detection algorithm (achieving recall of 94.9% and precision of 99.3%), the best architecture (achieving 86.79% accuracy) was integrated into a fully automated pipeline capable of analyzing extensive USV datasets with high reliability. Additionally, users can specify an individual minimum accuracy threshold based on their research needs. In this semi-automated setup, the pipeline selectively classifies calls with high pseudo-probability, leaving the rest for manual inspection. Our study focuses exclusively on neonatal USVs. As part of an ongoing phenotyping study, our pipeline has proven to be a valuable tool for identifying key differences in USVs produced by mice with autism-like behaviors.
Electrical Impedance Tomography: A Fair Comparative Study on Deep Learning and Analytic-based Approaches
Oct 28, 2023Abstract:Electrical Impedance Tomography (EIT) is a powerful imaging technique with diverse applications, e.g., medical diagnosis, industrial monitoring, and environmental studies. The EIT inverse problem is about inferring the internal conductivity distribution of an object from measurements taken on its boundary. It is severely ill-posed, necessitating advanced computational methods for accurate image reconstructions. Recent years have witnessed significant progress, driven by innovations in analytic-based approaches and deep learning. This review explores techniques for solving the EIT inverse problem, focusing on the interplay between contemporary deep learning-based strategies and classical analytic-based methods. Four state-of-the-art deep learning algorithms are rigorously examined, harnessing the representational capabilities of deep neural networks to reconstruct intricate conductivity distributions. In parallel, two analytic-based methods, rooted in mathematical formulations and regularisation techniques, are dissected for their strengths and limitations. These methodologies are evaluated through various numerical experiments, encompassing diverse scenarios that reflect real-world complexities. A suite of performance metrics is employed to assess the efficacy of these methods. These metrics collectively provide a nuanced understanding of the methods' ability to capture essential features and delineate complex conductivity patterns. One novel feature of the study is the incorporation of variable conductivity scenarios, introducing a level of heterogeneity that mimics textured inclusions. This departure from uniform conductivity assumptions mimics realistic scenarios where tissues or materials exhibit spatially varying electrical properties. Exploring how each method responds to such variable conductivity scenarios opens avenues for understanding their robustness and adaptability.
Steerable Conditional Diffusion for Out-of-Distribution Adaptation in Imaging Inverse Problems
Aug 28, 2023



Abstract:Denoising diffusion models have emerged as the go-to framework for solving inverse problems in imaging. A critical concern regarding these models is their performance on out-of-distribution (OOD) tasks, which remains an under-explored challenge. Realistic reconstructions inconsistent with the measured data can be generated, hallucinating image features that are uniquely present in the training dataset. To simultaneously enforce data-consistency and leverage data-driven priors, we introduce a novel sampling framework called Steerable Conditional Diffusion. This framework adapts the denoising network specifically to the available measured data. Utilising our proposed method, we achieve substantial enhancements in OOD performance across diverse imaging modalities, advancing the robust deployment of denoising diffusion models in real-world applications.
Score-Based Generative Models for PET Image Reconstruction
Aug 27, 2023



Abstract:Score-based generative models have demonstrated highly promising results for medical image reconstruction tasks in magnetic resonance imaging or computed tomography. However, their application to Positron Emission Tomography (PET) is still largely unexplored. PET image reconstruction involves a variety of challenges, including Poisson noise with high variance and a wide dynamic range. To address these challenges, we propose several PET-specific adaptations of score-based generative models. The proposed framework is developed for both 2D and 3D PET. In addition, we provide an extension to guided reconstruction using magnetic resonance images. We validate the approach through extensive 2D and 3D $\textit{in-silico}$ experiments with a model trained on patient-realistic data without lesions, and evaluate on data without lesions as well as out-of-distribution data with lesions. This demonstrates the proposed method's robustness and significant potential for improved PET reconstruction.
SVD-DIP: Overcoming the Overfitting Problem in DIP-based CT Reconstruction
Mar 28, 2023



Abstract:The deep image prior (DIP) is a well-established unsupervised deep learning method for image reconstruction; yet it is far from being flawless. The DIP overfits to noise if not early stopped, or optimized via a regularized objective. We build on the regularized fine-tuning of a pretrained DIP, by adopting a novel strategy that restricts the learning to the adaptation of singular values. The proposed SVD-DIP uses ad hoc convolutional layers whose pretrained parameters are decomposed via the singular value decomposition. Optimizing the DIP then solely consists in the fine-tuning of the singular values, while keeping the left and right singular vectors fixed. We thoroughly validate the proposed method on real-measured $\mu$CT data of a lotus root as well as two medical datasets (LoDoPaB and Mayo). We report significantly improved stability of the DIP optimization, by overcoming the overfitting to noise.
Model Stitching and Visualization How GAN Generators can Invert Networks in Real-Time
Feb 04, 2023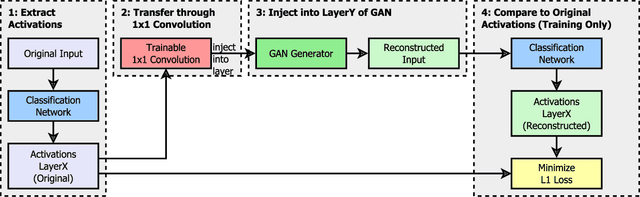
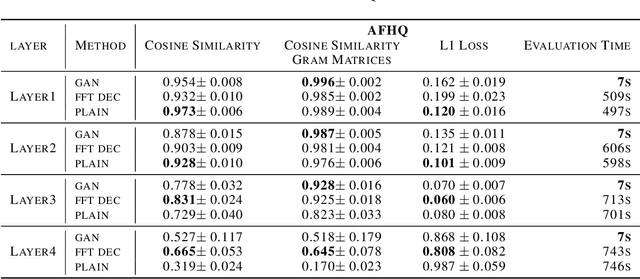
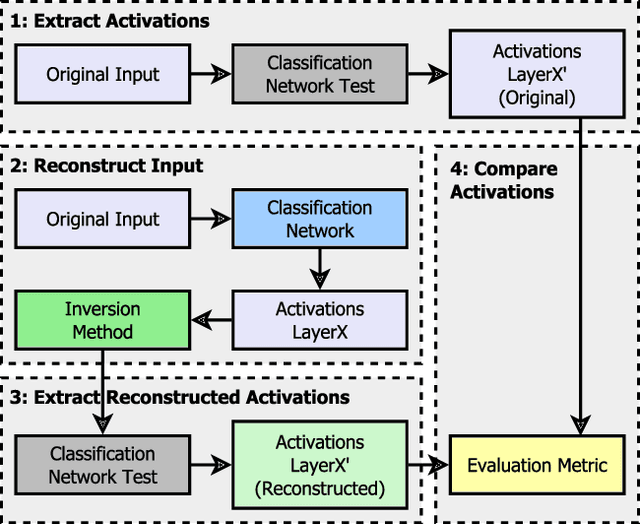
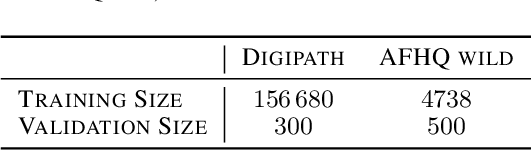
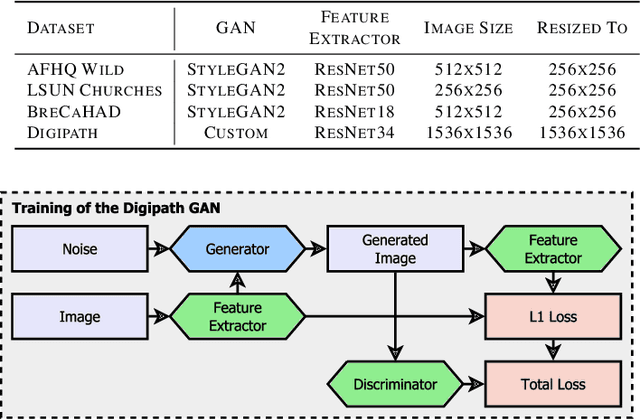
Abstract:Critical applications, such as in the medical field, require the rapid provision of additional information to interpret decisions made by deep learning methods. In this work, we propose a fast and accurate method to visualize activations of classification and semantic segmentation networks by stitching them with a GAN generator utilizing convolutions. We test our approach on images of animals from the AFHQ wild dataset and real-world digital pathology scans of stained tissue samples. Our method provides comparable results to established gradient descent methods on these datasets while running about two orders of magnitude faster.
Deep Learning Methods for Partial Differential Equations and Related Parameter Identification Problems
Dec 06, 2022Abstract:Recent years have witnessed a growth in mathematics for deep learning--which seeks a deeper understanding of the concepts of deep learning with mathematics, and explores how to make it more robust--and deep learning for mathematics, where deep learning algorithms are used to solve problems in mathematics. The latter has popularised the field of scientific machine learning where deep learning is applied to problems in scientific computing. Specifically, more and more neural network architectures have been developed to solve specific classes of partial differential equations (PDEs). Such methods exploit properties that are inherent to PDEs and thus solve the PDEs better than classical feed-forward neural networks, recurrent neural networks, and convolutional neural networks. This has had a great impact in the area of mathematical modeling where parametric PDEs are widely used to model most natural and physical processes arising in science and engineering, In this work, we review such methods and extend them for parametric studies as well as for solving the related inverse problems. We equally proceed to show their relevance in some industrial applications.
SELTO: Sample-Efficient Learned Topology Optimization
Sep 12, 2022



Abstract:We present a sample-efficient deep learning strategy for topology optimization. Our end-to-end approach is supervised and includes physics-based preprocessing and equivariant networks. We analyze how different components of our deep learning pipeline influence the number of required training samples via a large-scale comparison. The results demonstrate that including physical concepts not only drastically improves the sample efficiency but also the predictions' physical correctness. Finally, we publish two topology optimization datasets containing problems and corresponding ground truth solutions. We are confident that these datasets will improve comparability and future progress in the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge