Daniel Otero Baguer
University of Bremen, aisencia
Concept Based Explanations and Class Contrasting
Feb 05, 2025Abstract:Explaining deep neural networks is challenging, due to their large size and non-linearity. In this paper, we introduce a concept-based explanation method, in order to explain the prediction for an individual class, as well as contrasting any two classes, i.e. explain why the model predicts one class over the other. We test it on several openly available classification models trained on ImageNet1K, as well as on a segmentation model trained to detect tumor in stained tissue samples. We perform both qualitative and quantitative tests. For example, for a ResNet50 model from pytorch model zoo, we can use the explanation for why the model predicts a class 'A' to automatically select six dataset crops where the model does not predict class 'A'. The model then predicts class 'A' again for the newly combined image in 71\% of the cases (works for 710 out of the 1000 classes). The code including an .ipynb example is available on git: https://github.com/rherdt185/concept-based-explanations-and-class-contrasting.
Smooth Deep Saliency
Apr 04, 2024Abstract:In this work, we investigate methods to reduce the noise in deep saliency maps coming from convolutional downsampling, with the purpose of explaining how a deep learning model detects tumors in scanned histological tissue samples. Those methods make the investigated models more interpretable for gradient-based saliency maps, computed in hidden layers. We test our approach on different models trained for image classification on ImageNet1K, and models trained for tumor detection on Camelyon16 and in-house real-world digital pathology scans of stained tissue samples. Our results show that the checkerboard noise in the gradient gets reduced, resulting in smoother and therefore easier to interpret saliency maps.
Model Stitching and Visualization How GAN Generators can Invert Networks in Real-Time
Feb 04, 2023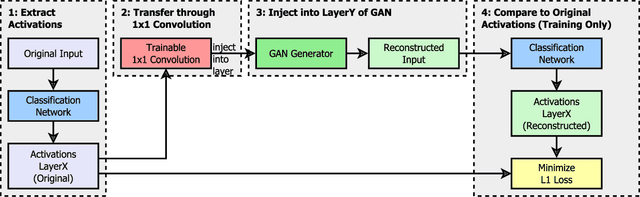
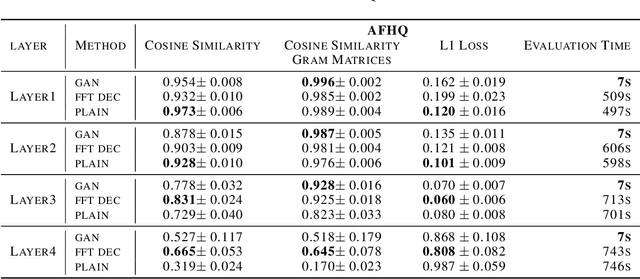
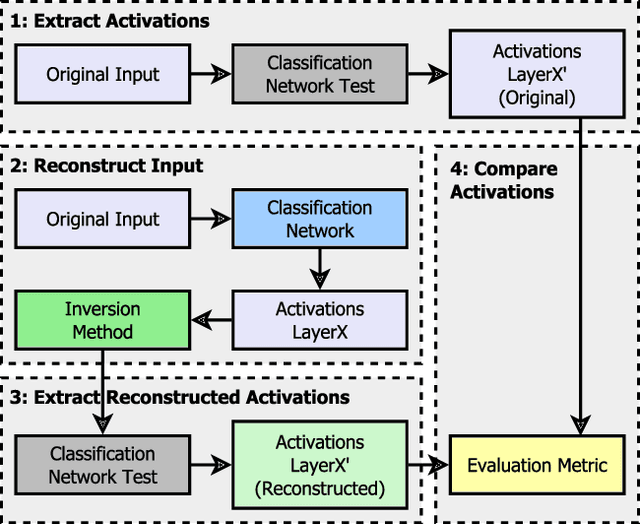
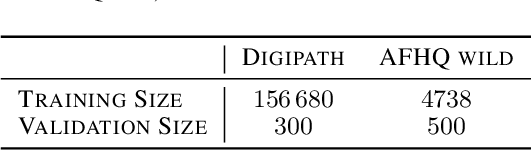
Abstract:Critical applications, such as in the medical field, require the rapid provision of additional information to interpret decisions made by deep learning methods. In this work, we propose a fast and accurate method to visualize activations of classification and semantic segmentation networks by stitching them with a GAN generator utilizing convolutions. We test our approach on images of animals from the AFHQ wild dataset and real-world digital pathology scans of stained tissue samples. Our method provides comparable results to established gradient descent methods on these datasets while running about two orders of magnitude faster.
Deeply supervised UNet for semantic segmentation to assist dermatopathological assessment of Basal Cell Carcinoma (BCC)
Mar 08, 2021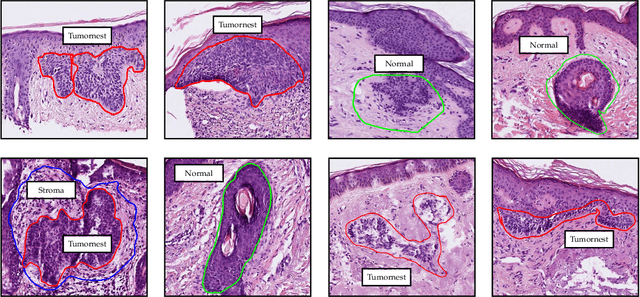
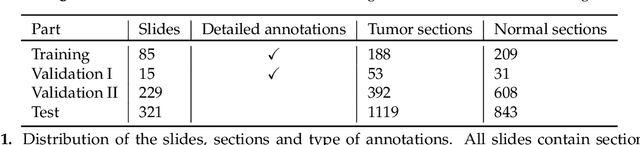
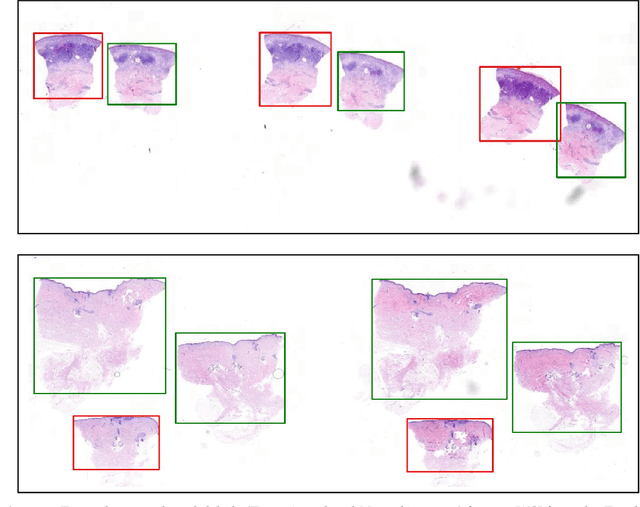
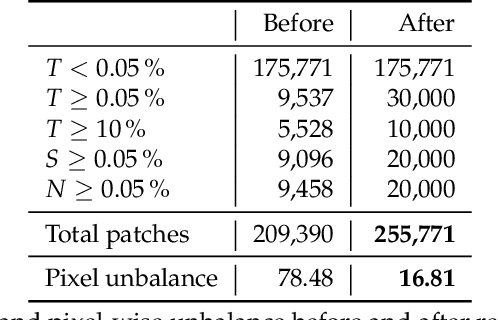
Abstract:Accurate and fast assessment of resection margins is an essential part of a dermatopathologist's clinical routine. In this work, we successfully develop a deep learning method to assist the pathologists by marking critical regions that have a high probability of exhibiting pathological features in Whole Slide Images (WSI). We focus on detecting Basal Cell Carcinoma (BCC) through semantic segmentation using several models based on the UNet architecture. The study includes 650 WSI with 3443 tissue sections in total. Two clinical dermatopathologists annotated the data, marking tumor tissues' exact location on 100 WSI. The rest of the data, with ground-truth section-wise labels, is used to further validate and test the models. We analyze two different encoders for the first part of the UNet network and two additional training strategies: a) deep supervision, b) linear combination of decoder outputs, and obtain some interpretations about what the network's decoder does in each case. The best model achieves over 96%, accuracy, sensitivity, and specificity on the test set.
Computed Tomography Reconstruction Using Deep Image Prior and Learned Reconstruction Methods
Mar 12, 2020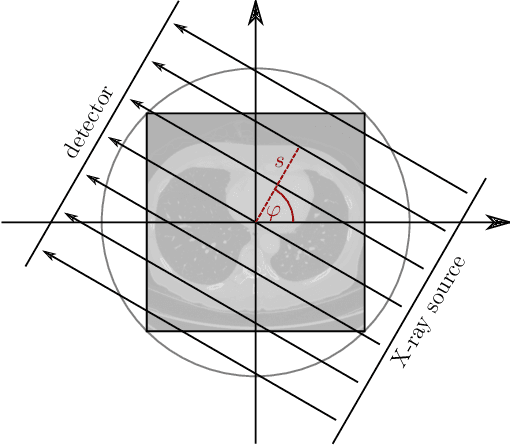
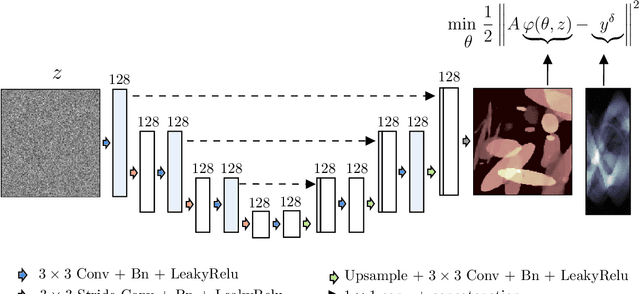

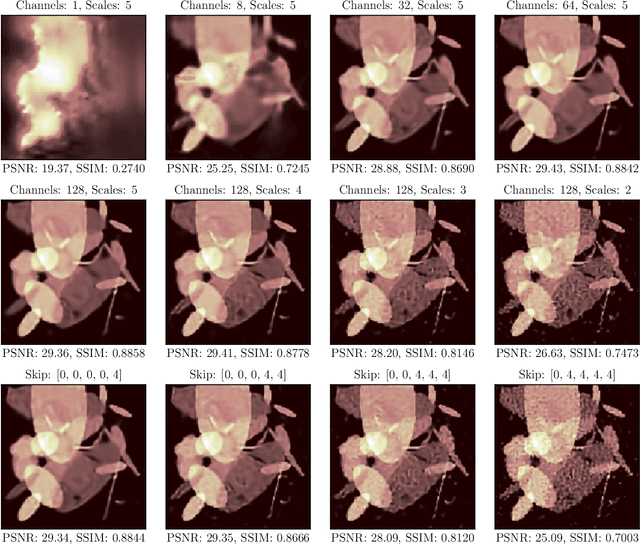
Abstract:In this work, we investigate the application of deep learning methods for computed tomography in the context of having a low-data regime. As motivation, we review some of the existing approaches and obtain quantitative results after training them with different amounts of data. We find that the learned primal-dual has an outstanding performance in terms of reconstruction quality and data efficiency. However, in general, end-to-end learned methods have two issues: a) lack of classical guarantees in inverse problems and b) lack of generalization when not trained with enough data. To overcome these issues, we bring in the deep image prior approach in combination with classical regularization. The proposed methods improve the state-of-the-art results in the low data-regime.
The LoDoPaB-CT Dataset: A Benchmark Dataset for Low-Dose CT Reconstruction Methods
Oct 01, 2019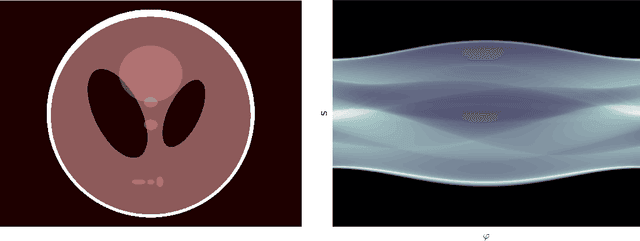

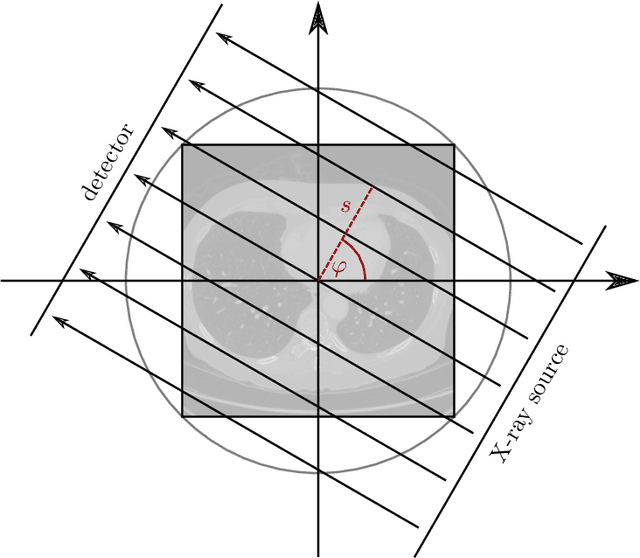
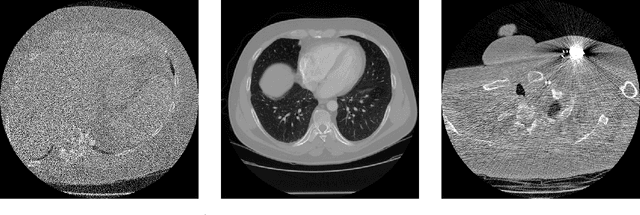
Abstract:Deep Learning approaches for solving Inverse Problems in imaging have become very effective and are demonstrated to be quite competitive in the field. Comparing these approaches is a challenging task since they highly rely on the data and the setup that is used for training. We provide a public dataset of computed tomography images and simulated low-dose measurements suitable for training this kind of methods. With the LoDoPaB-CT Dataset we aim to create a benchmark that allows for a fair comparison. It contains over 40,000 scan slices from around 800 patients selected from the LIDC/IDRI Database. In this paper we describe how we processed the original slices and how we simulated the measurements. We also include first baseline results.
Regularization by architecture: A deep prior approach for inverse problems
Dec 10, 2018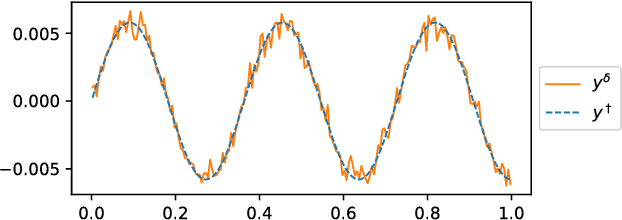
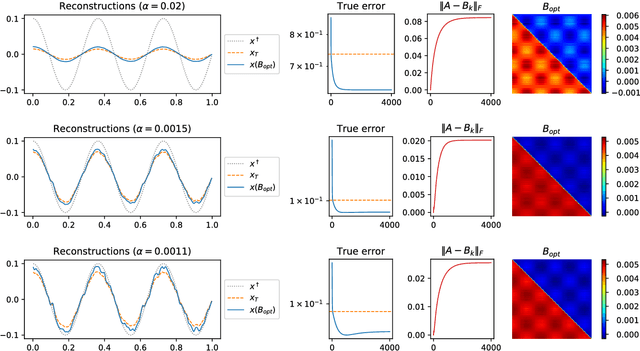
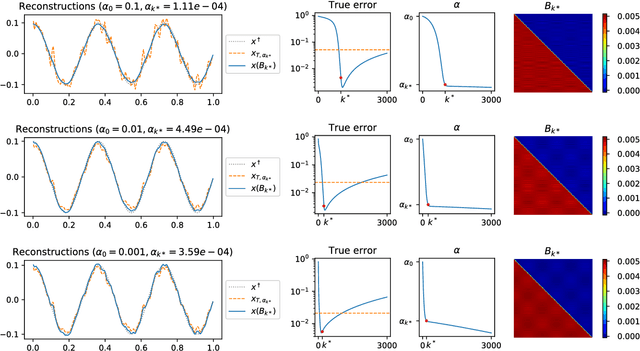
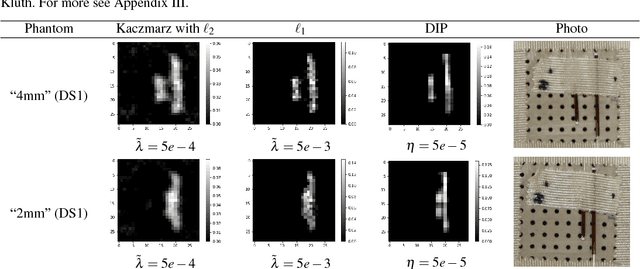
Abstract:The present paper studies the so called deep image prior (DIP) technique in the context of inverse problems. DIP networks have been introduced recently for applications in image processing, also first experimental results for applying DIP to inverse problems have been reported. This paper aims at discussing different interpretations of DIP and to obtain analytic results for specific network designs and linear operators. The main contribution is to introduce the idea of viewing these approaches as the optimization of Tiknonov functionals rather than optimizing networks. Besides theoretical results, we present numerical verifications for an academic example (integration operator) as well as for the inverse problem of magnetic particle imaging (MPI). The reconstructions obtained by deep prior networks are compared with state of the art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge