Tobias Boskamp
Deeply supervised UNet for semantic segmentation to assist dermatopathological assessment of Basal Cell Carcinoma (BCC)
Mar 08, 2021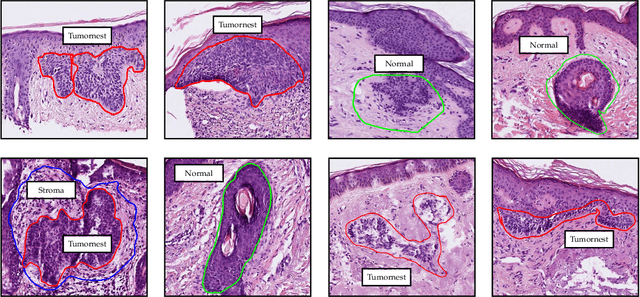
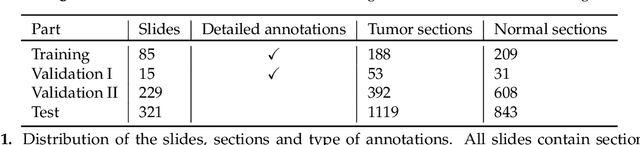
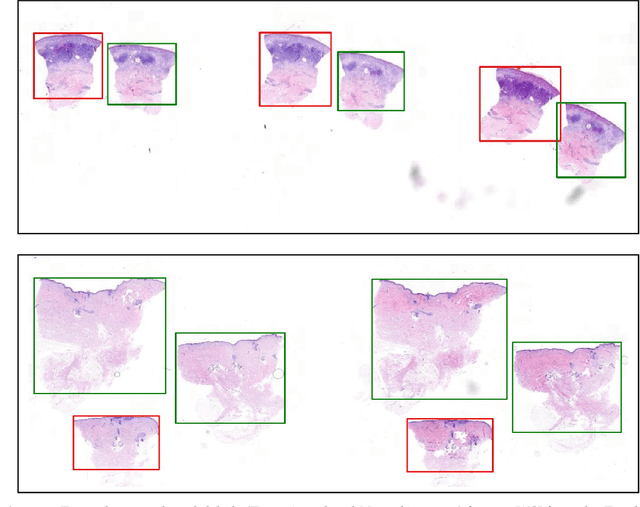
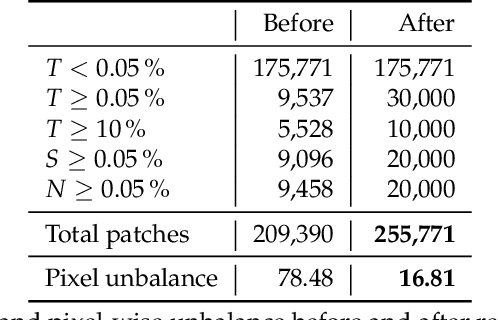
Abstract:Accurate and fast assessment of resection margins is an essential part of a dermatopathologist's clinical routine. In this work, we successfully develop a deep learning method to assist the pathologists by marking critical regions that have a high probability of exhibiting pathological features in Whole Slide Images (WSI). We focus on detecting Basal Cell Carcinoma (BCC) through semantic segmentation using several models based on the UNet architecture. The study includes 650 WSI with 3443 tissue sections in total. Two clinical dermatopathologists annotated the data, marking tumor tissues' exact location on 100 WSI. The rest of the data, with ground-truth section-wise labels, is used to further validate and test the models. We analyze two different encoders for the first part of the UNet network and two additional training strategies: a) deep supervision, b) linear combination of decoder outputs, and obtain some interpretations about what the network's decoder does in each case. The best model achieves over 96%, accuracy, sensitivity, and specificity on the test set.
Deep Relevance Regularization: Interpretable and Robust Tumor Typing of Imaging Mass Spectrometry Data
Dec 10, 2019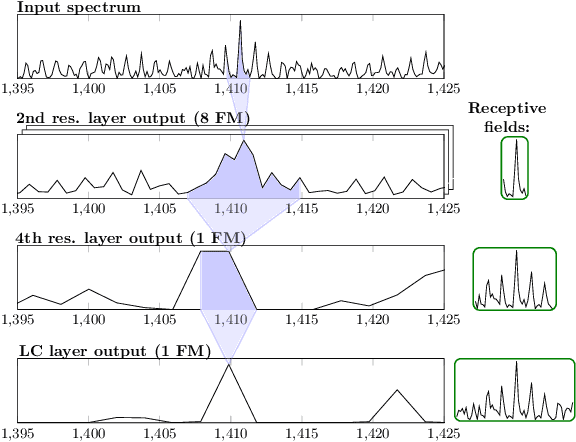
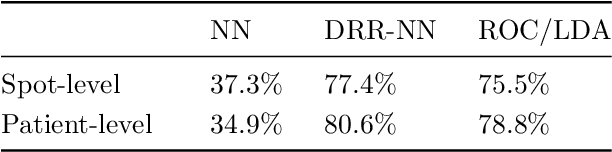
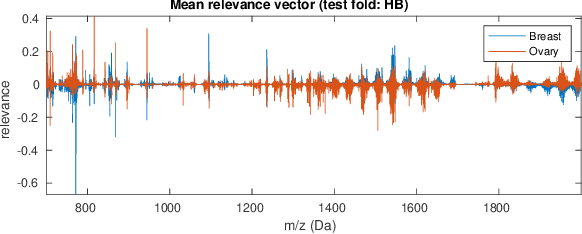
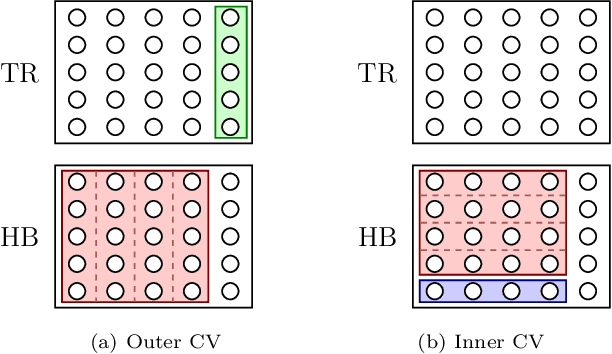
Abstract:Neural networks have recently been established as a viable classification method for imaging mass spectrometry data for tumor typing. For multi-laboratory scenarios however, certain confounding factors may strongly impede their performance. In this work, we introduce Deep Relevance Regularization, a method of restricting what the neural network can focus on during classification, in order to improve the classification performance. We demonstrate how Deep Relevance Regularization robustifies neural networks against confounding factors on a challenging inter-lab dataset consisting of breast and ovarian carcinoma. We further show that this makes the relevance map -- a way of visualizing the discriminative parts of the mass spectrum -- sparser, thereby making the classifier easier to interpret
Deep Learning for Tumor Classification in Imaging Mass Spectrometry
Oct 20, 2017
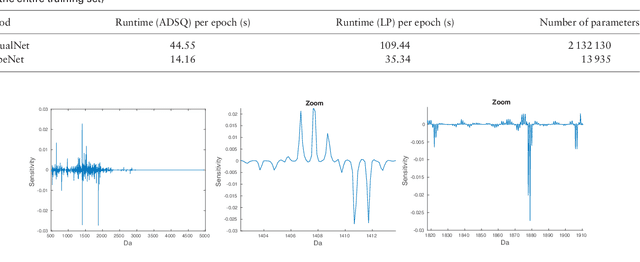
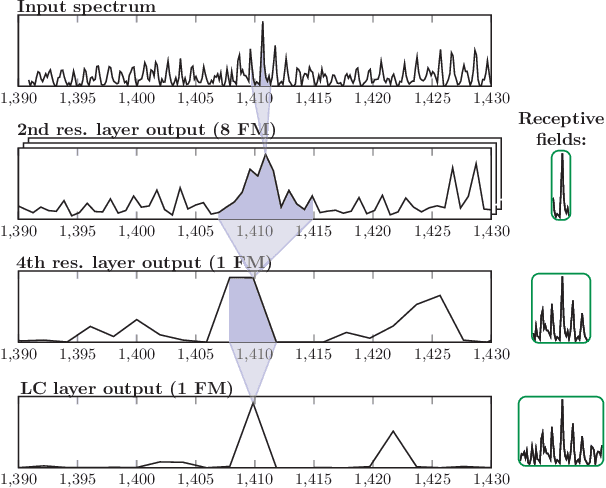

Abstract:Motivation: Tumor classification using Imaging Mass Spectrometry (IMS) data has a high potential for future applications in pathology. Due to the complexity and size of the data, automated feature extraction and classification steps are required to fully process the data. Deep learning offers an approach to learn feature extraction and classification combined in a single model. Commonly these steps are handled separately in IMS data analysis, hence deep learning offers an alternative strategy worthwhile to explore. Results: Methodologically, we propose an adapted architecture based on deep convolutional networks to handle the characteristics of mass spectrometry data, as well as a strategy to interpret the learned model in the spectral domain based on a sensitivity analysis. The proposed methods are evaluated on two challenging tumor classification tasks and compared to a baseline approach. Competitiveness of the proposed methods are shown on both tasks by studying the performance via cross-validation. Moreover, the learned models are analyzed by the proposed sensitivity analysis revealing biologically plausible effects as well as confounding factors of the considered task. Thus, this study may serve as a starting point for further development of deep learning approaches in IMS classification tasks.
* 10 pages, 5 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge