Deep Learning for Tumor Classification in Imaging Mass Spectrometry
Paper and Code
Oct 20, 2017
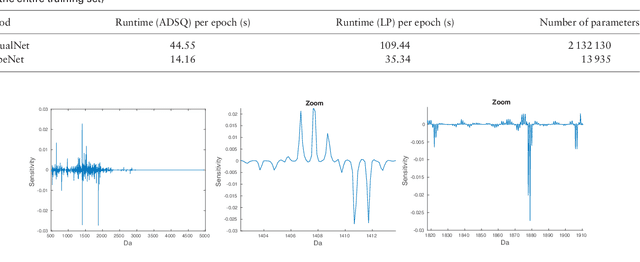
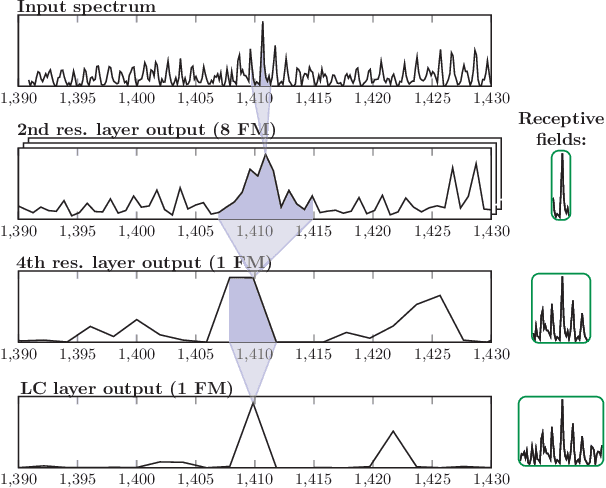

Motivation: Tumor classification using Imaging Mass Spectrometry (IMS) data has a high potential for future applications in pathology. Due to the complexity and size of the data, automated feature extraction and classification steps are required to fully process the data. Deep learning offers an approach to learn feature extraction and classification combined in a single model. Commonly these steps are handled separately in IMS data analysis, hence deep learning offers an alternative strategy worthwhile to explore. Results: Methodologically, we propose an adapted architecture based on deep convolutional networks to handle the characteristics of mass spectrometry data, as well as a strategy to interpret the learned model in the spectral domain based on a sensitivity analysis. The proposed methods are evaluated on two challenging tumor classification tasks and compared to a baseline approach. Competitiveness of the proposed methods are shown on both tasks by studying the performance via cross-validation. Moreover, the learned models are analyzed by the proposed sensitivity analysis revealing biologically plausible effects as well as confounding factors of the considered task. Thus, this study may serve as a starting point for further development of deep learning approaches in IMS classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge