Bruce Rosen
DeepNeuro: an open-source deep learning toolbox for neuroimaging
Aug 14, 2018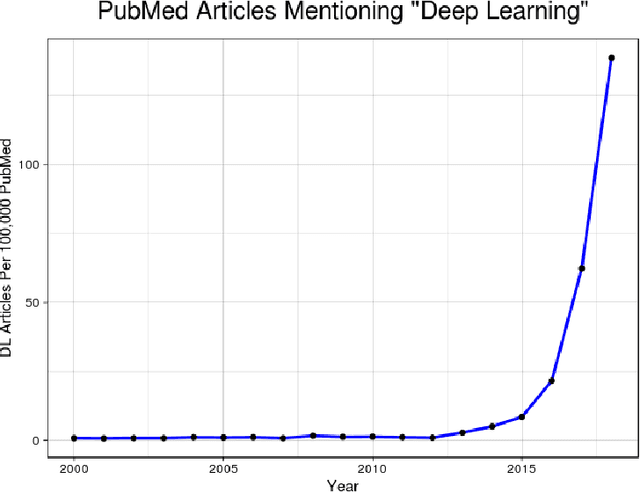
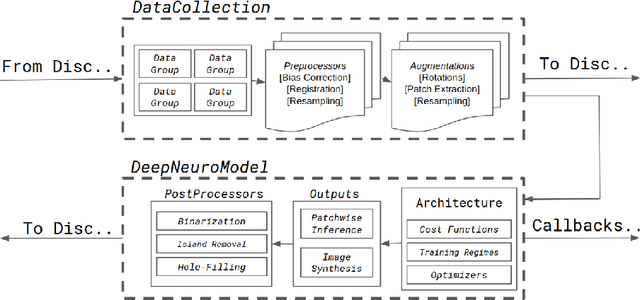
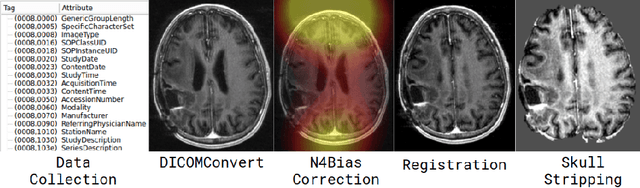
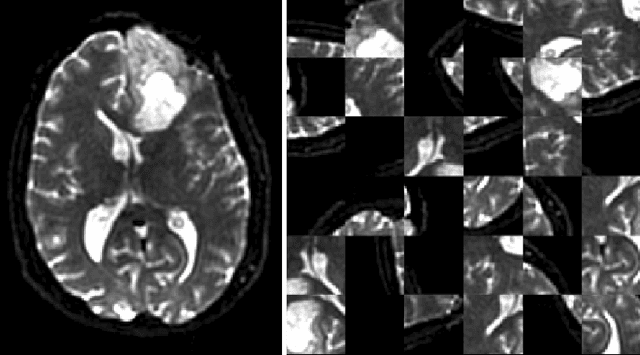
Abstract:Translating neural networks from theory to clinical practice has unique challenges, specifically in the field of neuroimaging. In this paper, we present DeepNeuro, a deep learning framework that is best-suited to putting deep learning algorithms for neuroimaging in practical usage with a minimum of friction. We show how this framework can be used to both design and train neural network architectures, as well as modify state-of-the-art architectures in a flexible and intuitive way. We display the pre- and postprocessing functions common in the medical imaging community that DeepNeuro offers to ensure consistent performance of networks across variable users, institutions, and scanners. And we show how pipelines created in DeepNeuro can be concisely packaged into shareable Docker containers and command-line interfaces using DeepNeuro's pipeline resources.
Sequential 3D U-Nets for Biologically-Informed Brain Tumor Segmentation
Sep 09, 2017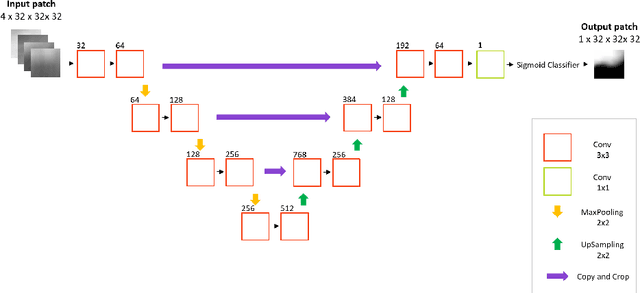

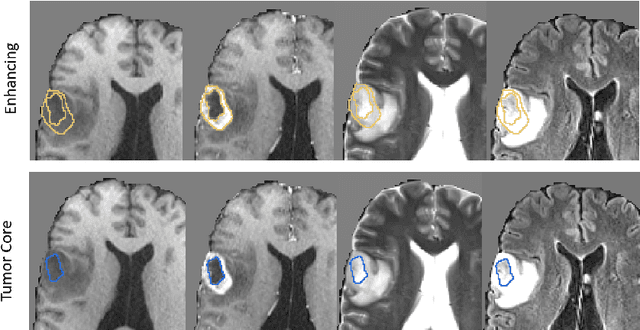
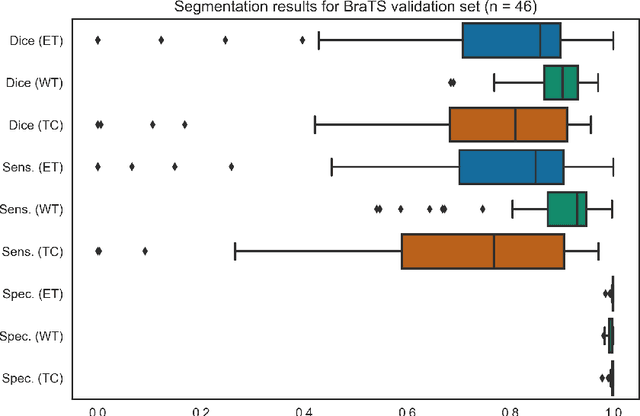
Abstract:Deep learning has quickly become the weapon of choice for brain lesion segmentation. However, few existing algorithms pre-configure any biological context of their chosen segmentation tissues, and instead rely on the neural network's optimizer to develop such associations de novo. We present a novel method for applying deep neural networks to the problem of glioma tissue segmentation that takes into account the structured nature of gliomas - edematous tissue surrounding mutually-exclusive regions of enhancing and non-enhancing tumor. We trained multiple deep neural networks with a 3D U-Net architecture in a tree structure to create segmentations for edema, non-enhancing tumor, and enhancing tumor regions. Specifically, training was configured such that the whole tumor region including edema was predicted first, and its output segmentation was fed as input into separate models to predict enhancing and non-enhancing tumor. Our method was trained and evaluated on the publicly available BraTS dataset, achieving Dice scores of 0.882, 0.732, and 0.730 for whole tumor, enhancing tumor and tumor core respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge