João Matos
Classification of Keratitis from Eye Corneal Photographs using Deep Learning
Nov 13, 2024



Abstract:Keratitis is an inflammatory corneal condition responsible for 10% of visual impairment in low- and middle-income countries (LMICs), with bacteria, fungi, or amoeba as the most common infection etiologies. While an accurate and timely diagnosis is crucial for the selected treatment and the patients' sight outcomes, due to the high cost and limited availability of laboratory diagnostics in LMICs, diagnosis is often made by clinical observation alone, despite its lower accuracy. In this study, we investigate and compare different deep learning approaches to diagnose the source of infection: 1) three separate binary models for infection type predictions; 2) a multitask model with a shared backbone and three parallel classification layers (Multitask V1); and, 3) a multitask model with a shared backbone and a multi-head classification layer (Multitask V2). We used a private Brazilian cornea dataset to conduct the empirical evaluation. We achieved the best results with Multitask V2, with an area under the receiver operating characteristic curve (AUROC) confidence intervals of 0.7413-0.7740 (bacteria), 0.8395-0.8725 (fungi), and 0.9448-0.9616 (amoeba). A statistical analysis of the impact of patient features on models' performance revealed that sex significantly affects amoeba infection prediction, and age seems to affect fungi and bacteria predictions.
WorldMedQA-V: a multilingual, multimodal medical examination dataset for multimodal language models evaluation
Oct 16, 2024Abstract:Multimodal/vision language models (VLMs) are increasingly being deployed in healthcare settings worldwide, necessitating robust benchmarks to ensure their safety, efficacy, and fairness. Multiple-choice question and answer (QA) datasets derived from national medical examinations have long served as valuable evaluation tools, but existing datasets are largely text-only and available in a limited subset of languages and countries. To address these challenges, we present WorldMedQA-V, an updated multilingual, multimodal benchmarking dataset designed to evaluate VLMs in healthcare. WorldMedQA-V includes 568 labeled multiple-choice QAs paired with 568 medical images from four countries (Brazil, Israel, Japan, and Spain), covering original languages and validated English translations by native clinicians, respectively. Baseline performance for common open- and closed-source models are provided in the local language and English translations, and with and without images provided to the model. The WorldMedQA-V benchmark aims to better match AI systems to the diverse healthcare environments in which they are deployed, fostering more equitable, effective, and representative applications.
Evaluating the Impact of Pulse Oximetry Bias in Machine Learning under Counterfactual Thinking
Aug 08, 2024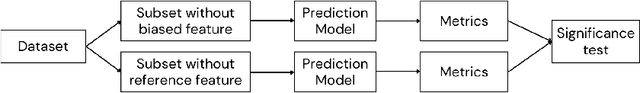
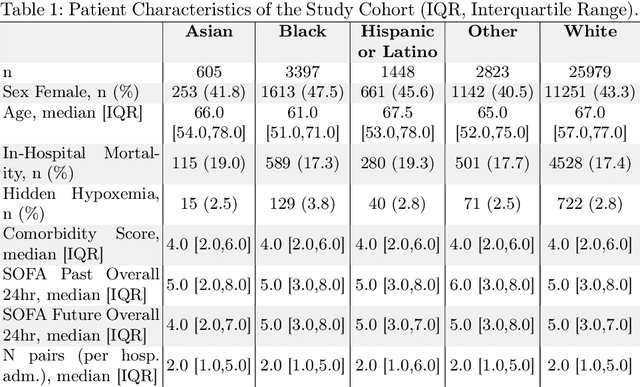
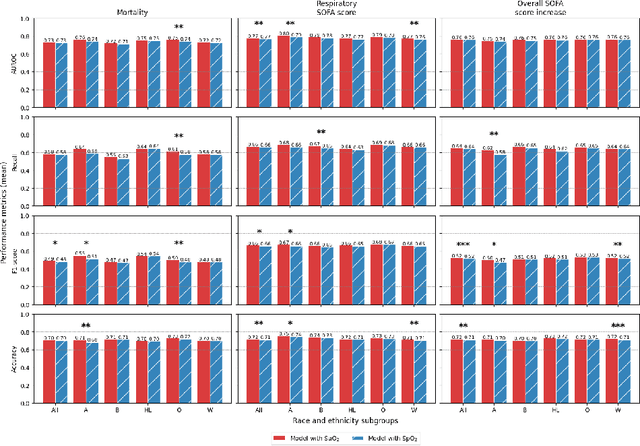

Abstract:Algorithmic bias in healthcare mirrors existing data biases. However, the factors driving unfairness are not always known. Medical devices capture significant amounts of data but are prone to errors; for instance, pulse oximeters overestimate the arterial oxygen saturation of darker-skinned individuals, leading to worse outcomes. The impact of this bias in machine learning (ML) models remains unclear. This study addresses the technical challenges of quantifying the impact of medical device bias in downstream ML. Our experiments compare a "perfect world", without pulse oximetry bias, using SaO2 (blood-gas), to the "actual world", with biased measurements, using SpO2 (pulse oximetry). Under this counterfactual design, two models are trained with identical data, features, and settings, except for the method of measuring oxygen saturation: models using SaO2 are a "control" and models using SpO2 a "treatment". The blood-gas oximetry linked dataset was a suitable test-bed, containing 163,396 nearly-simultaneous SpO2 - SaO2 paired measurements, aligned with a wide array of clinical features and outcomes. We studied three classification tasks: in-hospital mortality, respiratory SOFA score in the next 24 hours, and SOFA score increase by two points. Models using SaO2 instead of SpO2 generally showed better performance. Patients with overestimation of O2 by pulse oximetry of > 3% had significant decreases in mortality prediction recall, from 0.63 to 0.59, P < 0.001. This mirrors clinical processes where biased pulse oximetry readings provide clinicians with false reassurance of patients' oxygen levels. A similar degradation happened in ML models, with pulse oximetry biases leading to more false negatives in predicting adverse outcomes.
EHRmonize: A Framework for Medical Concept Abstraction from Electronic Health Records using Large Language Models
Jun 28, 2024



Abstract:Electronic health records (EHRs) contain vast amounts of complex data, but harmonizing and processing this information remains a challenging and costly task requiring significant clinical expertise. While large language models (LLMs) have shown promise in various healthcare applications, their potential for abstracting medical concepts from EHRs remains largely unexplored. We introduce EHRmonize, a framework leveraging LLMs to abstract medical concepts from EHR data. Our study uses medication data from two real-world EHR databases to evaluate five LLMs on two free-text extraction and six binary classification tasks across various prompting strategies. GPT-4o's with 10-shot prompting achieved the highest performance in all tasks, accompanied by Claude-3.5-Sonnet in a subset of tasks. GPT-4o achieved an accuracy of 97% in identifying generic route names, 82% for generic drug names, and 100% in performing binary classification of antibiotics. While EHRmonize significantly enhances efficiency, reducing annotation time by an estimated 60%, we emphasize that clinician oversight remains essential. Our framework, available as a Python package, offers a promising tool to assist clinicians in EHR data abstraction, potentially accelerating healthcare research and improving data harmonization processes.
Analyzing Diversity in Healthcare LLM Research: A Scientometric Perspective
Jun 19, 2024Abstract:The deployment of large language models (LLMs) in healthcare has demonstrated substantial potential for enhancing clinical decision-making, administrative efficiency, and patient outcomes. However, the underrepresentation of diverse groups in the development and application of these models can perpetuate biases, leading to inequitable healthcare delivery. This paper presents a comprehensive scientometric analysis of LLM research for healthcare, including data from January 1, 2021, to June 16, 2024. By analyzing metadata from PubMed and Dimensions, including author affiliations, countries, and funding sources, we assess the diversity of contributors to LLM research. Our findings highlight significant gender and geographic disparities, with a predominance of male authors and contributions primarily from high-income countries (HICs). We introduce a novel journal diversity index based on Gini impurity to measure the inclusiveness of scientific publications. Our results underscore the necessity for greater representation in order to ensure the equitable application of LLMs in healthcare. We propose actionable strategies to enhance diversity and inclusivity in artificial intelligence research, with the ultimate goal of fostering a more inclusive and equitable future in healthcare innovation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge