Richard Brown
DeepEdit: Deep Editable Learning for Interactive Segmentation of 3D Medical Images
May 18, 2023Abstract:Automatic segmentation of medical images is a key step for diagnostic and interventional tasks. However, achieving this requires large amounts of annotated volumes, which can be tedious and time-consuming task for expert annotators. In this paper, we introduce DeepEdit, a deep learning-based method for volumetric medical image annotation, that allows automatic and semi-automatic segmentation, and click-based refinement. DeepEdit combines the power of two methods: a non-interactive (i.e. automatic segmentation using nnU-Net, UNET or UNETR) and an interactive segmentation method (i.e. DeepGrow), into a single deep learning model. It allows easy integration of uncertainty-based ranking strategies (i.e. aleatoric and epistemic uncertainty computation) and active learning. We propose and implement a method for training DeepEdit by using standard training combined with user interaction simulation. Once trained, DeepEdit allows clinicians to quickly segment their datasets by using the algorithm in auto segmentation mode or by providing clicks via a user interface (i.e. 3D Slicer, OHIF). We show the value of DeepEdit through evaluation on the PROSTATEx dataset for prostate/prostatic lesions and the Multi-Atlas Labeling Beyond the Cranial Vault (BTCV) dataset for abdominal CT segmentation, using state-of-the-art network architectures as baseline for comparison. DeepEdit could reduce the time and effort annotating 3D medical images compared to DeepGrow alone. Source code is available at https://github.com/Project-MONAI/MONAILabel
MONAI: An open-source framework for deep learning in healthcare
Nov 04, 2022



Abstract:Artificial Intelligence (AI) is having a tremendous impact across most areas of science. Applications of AI in healthcare have the potential to improve our ability to detect, diagnose, prognose, and intervene on human disease. For AI models to be used clinically, they need to be made safe, reproducible and robust, and the underlying software framework must be aware of the particularities (e.g. geometry, physiology, physics) of medical data being processed. This work introduces MONAI, a freely available, community-supported, and consortium-led PyTorch-based framework for deep learning in healthcare. MONAI extends PyTorch to support medical data, with a particular focus on imaging, and provide purpose-specific AI model architectures, transformations and utilities that streamline the development and deployment of medical AI models. MONAI follows best practices for software-development, providing an easy-to-use, robust, well-documented, and well-tested software framework. MONAI preserves the simple, additive, and compositional approach of its underlying PyTorch libraries. MONAI is being used by and receiving contributions from research, clinical and industrial teams from around the world, who are pursuing applications spanning nearly every aspect of healthcare.
Metadata-Based Detection of Child Sexual Abuse Material
Oct 05, 2020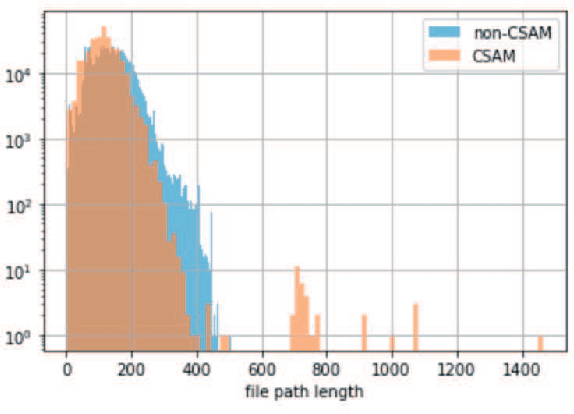
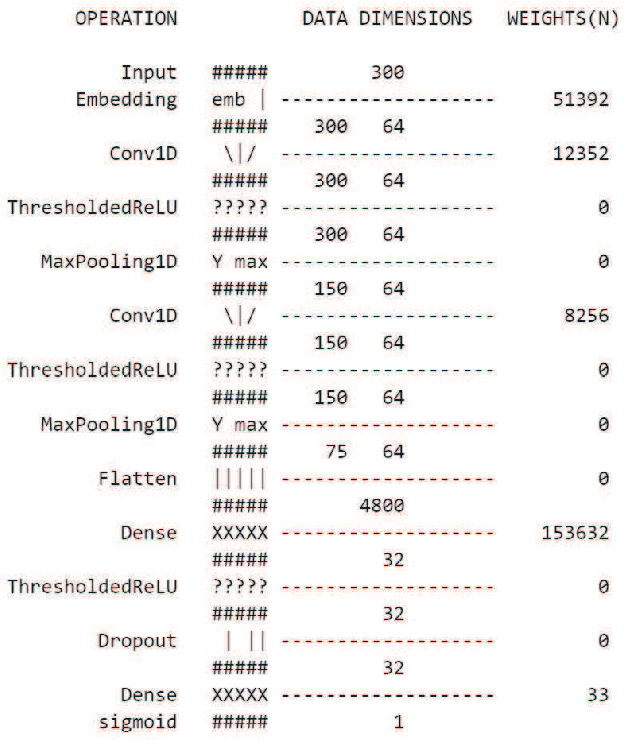
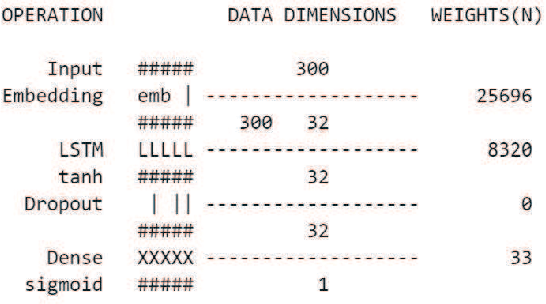
Abstract:In the last decade, the scale of creation and distribution of child sexual abuse medias (CSAM) has exponentially increased. Technologies that aid law enforcement agencies worldwide to identify such crimes rapidly can potentially result in the mitigation of child victimization, and the apprehending of offenders. Machine learning presents the potential to help law enforcement rapidly identify such material, and even block such content from being distributed digitally. However, collecting and storing CSAM files to train machine learning models has many ethical and legal constraints, creating a barrier to the development of accurate computer vision-based models. With such restrictions in place, the development of accurate machine learning classifiers for CSAM identification based on file metadata becomes crucial. In this work, we propose a system for CSAM identification on file storage systems based solely on metadata - file paths. Our aim is to provide a tool that is material type agnostic (image, video, PDF), and can potentially scans thousands of file storage systems in a short time. Our approach uses convolutional neural networks, and achieves an accuracy of 97% and recall of 94%. Additionally, we address the potential problem of offenders trying to evade detection by this model by evaluating the robustness of our model against adversarial modifications in the file paths.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge