Andres Diaz-Pinto
In search of truth: Evaluating concordance of AI-based anatomy segmentation models
Dec 17, 2025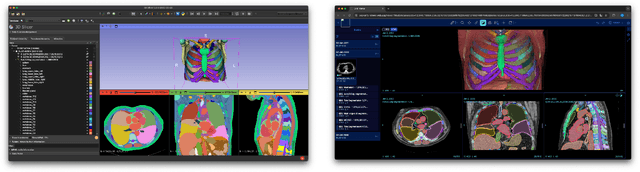


Abstract:Purpose AI-based methods for anatomy segmentation can help automate characterization of large imaging datasets. The growing number of similar in functionality models raises the challenge of evaluating them on datasets that do not contain ground truth annotations. We introduce a practical framework to assist in this task. Approach We harmonize the segmentation results into a standard, interoperable representation, which enables consistent, terminology-based labeling of the structures. We extend 3D Slicer to streamline loading and comparison of these harmonized segmentations, and demonstrate how standard representation simplifies review of the results using interactive summary plots and browser-based visualization using OHIF Viewer. To demonstrate the utility of the approach we apply it to evaluating segmentation of 31 anatomical structures (lungs, vertebrae, ribs, and heart) by six open-source models - TotalSegmentator 1.5 and 2.6, Auto3DSeg, MOOSE, MultiTalent, and CADS - for a sample of Computed Tomography (CT) scans from the publicly available National Lung Screening Trial (NLST) dataset. Results We demonstrate the utility of the framework in enabling automating loading, structure-wise inspection and comparison across models. Preliminary results ascertain practical utility of the approach in allowing quick detection and review of problematic results. The comparison shows excellent agreement segmenting some (e.g., lung) but not all structures (e.g., some models produce invalid vertebrae or rib segmentations). Conclusions The resources developed are linked from https://imagingdatacommons.github.io/segmentation-comparison/ including segmentation harmonization scripts, summary plots, and visualization tools. This work assists in model evaluation in absence of ground truth, ultimately enabling informed model selection.
SuFIA-BC: Generating High Quality Demonstration Data for Visuomotor Policy Learning in Surgical Subtasks
Apr 21, 2025



Abstract:Behavior cloning facilitates the learning of dexterous manipulation skills, yet the complexity of surgical environments, the difficulty and expense of obtaining patient data, and robot calibration errors present unique challenges for surgical robot learning. We provide an enhanced surgical digital twin with photorealistic human anatomical organs, integrated into a comprehensive simulator designed to generate high-quality synthetic data to solve fundamental tasks in surgical autonomy. We present SuFIA-BC: visual Behavior Cloning policies for Surgical First Interactive Autonomy Assistants. We investigate visual observation spaces including multi-view cameras and 3D visual representations extracted from a single endoscopic camera view. Through systematic evaluation, we find that the diverse set of photorealistic surgical tasks introduced in this work enables a comprehensive evaluation of prospective behavior cloning models for the unique challenges posed by surgical environments. We observe that current state-of-the-art behavior cloning techniques struggle to solve the contact-rich and complex tasks evaluated in this work, regardless of their underlying perception or control architectures. These findings highlight the importance of customizing perception pipelines and control architectures, as well as curating larger-scale synthetic datasets that meet the specific demands of surgical tasks. Project website: https://orbit-surgical.github.io/sufia-bc/
Segment Any Medical Model Extended
Mar 26, 2024Abstract:The Segment Anything Model (SAM) has drawn significant attention from researchers who work on medical image segmentation because of its generalizability. However, researchers have found that SAM may have limited performance on medical images compared to state-of-the-art non-foundation models. Regardless, the community sees potential in extending, fine-tuning, modifying, and evaluating SAM for analysis of medical imaging. An increasing number of works have been published focusing on the mentioned four directions, where variants of SAM are proposed. To this end, a unified platform helps push the boundary of the foundation model for medical images, facilitating the use, modification, and validation of SAM and its variants in medical image segmentation. In this work, we introduce SAMM Extended (SAMME), a platform that integrates new SAM variant models, adopts faster communication protocols, accommodates new interactive modes, and allows for fine-tuning of subcomponents of the models. These features can expand the potential of foundation models like SAM, and the results can be translated to applications such as image-guided therapy, mixed reality interaction, robotic navigation, and data augmentation.
DeepEdit: Deep Editable Learning for Interactive Segmentation of 3D Medical Images
May 18, 2023Abstract:Automatic segmentation of medical images is a key step for diagnostic and interventional tasks. However, achieving this requires large amounts of annotated volumes, which can be tedious and time-consuming task for expert annotators. In this paper, we introduce DeepEdit, a deep learning-based method for volumetric medical image annotation, that allows automatic and semi-automatic segmentation, and click-based refinement. DeepEdit combines the power of two methods: a non-interactive (i.e. automatic segmentation using nnU-Net, UNET or UNETR) and an interactive segmentation method (i.e. DeepGrow), into a single deep learning model. It allows easy integration of uncertainty-based ranking strategies (i.e. aleatoric and epistemic uncertainty computation) and active learning. We propose and implement a method for training DeepEdit by using standard training combined with user interaction simulation. Once trained, DeepEdit allows clinicians to quickly segment their datasets by using the algorithm in auto segmentation mode or by providing clicks via a user interface (i.e. 3D Slicer, OHIF). We show the value of DeepEdit through evaluation on the PROSTATEx dataset for prostate/prostatic lesions and the Multi-Atlas Labeling Beyond the Cranial Vault (BTCV) dataset for abdominal CT segmentation, using state-of-the-art network architectures as baseline for comparison. DeepEdit could reduce the time and effort annotating 3D medical images compared to DeepGrow alone. Source code is available at https://github.com/Project-MONAI/MONAILabel
MONAI: An open-source framework for deep learning in healthcare
Nov 04, 2022



Abstract:Artificial Intelligence (AI) is having a tremendous impact across most areas of science. Applications of AI in healthcare have the potential to improve our ability to detect, diagnose, prognose, and intervene on human disease. For AI models to be used clinically, they need to be made safe, reproducible and robust, and the underlying software framework must be aware of the particularities (e.g. geometry, physiology, physics) of medical data being processed. This work introduces MONAI, a freely available, community-supported, and consortium-led PyTorch-based framework for deep learning in healthcare. MONAI extends PyTorch to support medical data, with a particular focus on imaging, and provide purpose-specific AI model architectures, transformations and utilities that streamline the development and deployment of medical AI models. MONAI follows best practices for software-development, providing an easy-to-use, robust, well-documented, and well-tested software framework. MONAI preserves the simple, additive, and compositional approach of its underlying PyTorch libraries. MONAI is being used by and receiving contributions from research, clinical and industrial teams from around the world, who are pursuing applications spanning nearly every aspect of healthcare.
MONAI Label: A framework for AI-assisted Interactive Labeling of 3D Medical Images
Mar 23, 2022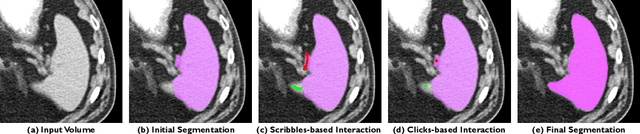
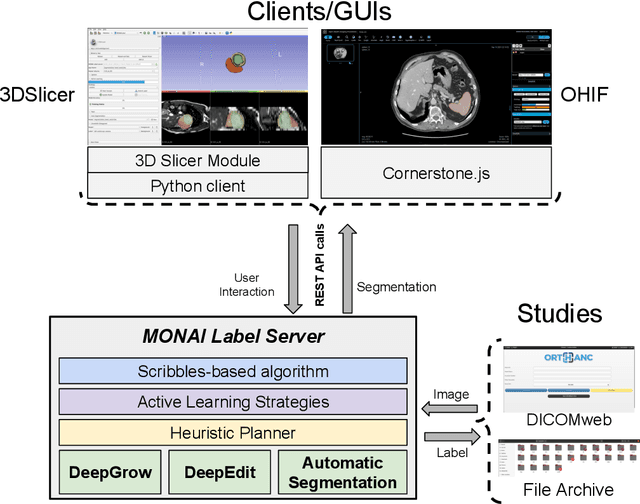
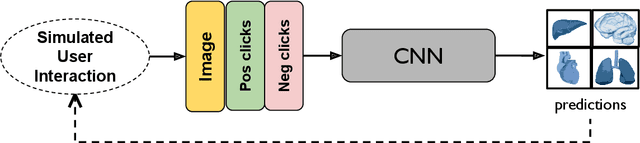
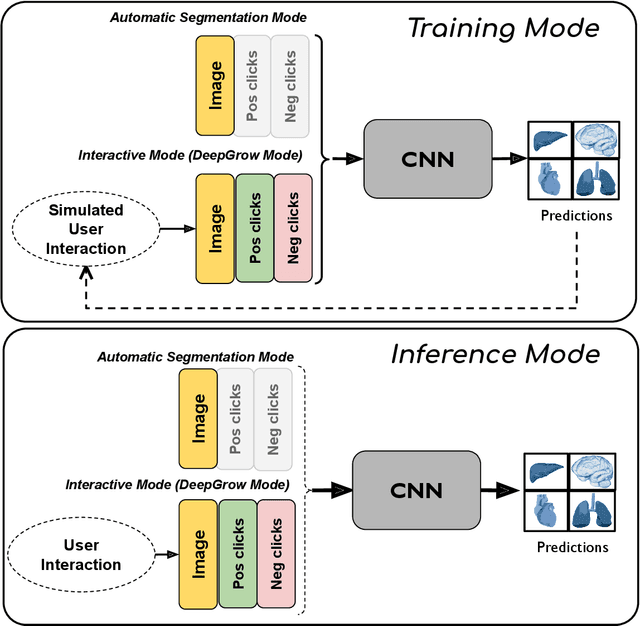
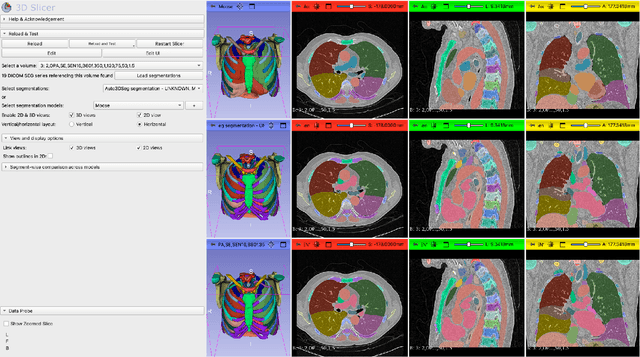
Abstract:The lack of annotated datasets is a major challenge in training new task-specific supervised AI algorithms as manual annotation is expensive and time-consuming. To address this problem, we present MONAI Label, a free and open-source platform that facilitates the development of AI-based applications that aim at reducing the time required to annotate 3D medical image datasets. Through MONAI Label researchers can develop annotation applications focusing on their domain of expertise. It allows researchers to readily deploy their apps as services, which can be made available to clinicians via their preferred user-interface. Currently, MONAI Label readily supports locally installed (3DSlicer) and web-based (OHIF) frontends, and offers two Active learning strategies to facilitate and speed up the training of segmentation algorithms. MONAI Label allows researchers to make incremental improvements to their labeling apps by making them available to other researchers and clinicians alike. Lastly, MONAI Label provides sample labeling apps, namely DeepEdit and DeepGrow, demonstrating dramatically reduced annotation times.
Semi-Automatic Algorithm for Breast MRI Lesion Segmentation Using Marker-Controlled Watershed Transformation
Dec 14, 2017


Abstract:Magnetic resonance imaging (MRI) is an effective imaging modality for identifying and localizing breast lesions in women. Accurate and precise lesion segmentation using a computer-aided-diagnosis (CAD) system, is a crucial step in evaluating tumor volume and in the quantification of tumor characteristics. However, this is a challenging task, since breast lesions have sophisticated shape, topological structure, and high variance in their intensity distribution across patients. In this paper, we propose a novel marker-controlled watershed transformation-based approach, which uses the brightest pixels in a region of interest (determined by experts) as markers to overcome this challenge, and accurately segment lesions in breast MRI. The proposed approach was evaluated on 106 lesions, which includes 64 malignant and 42 benign cases. Segmentation results were quantified by comparison with ground truth labels, using the Dice similarity coefficient (DSC) and Jaccard index (JI) metrics. The proposed method achieved an average Dice coefficient of 0.7808$\pm$0.1729 and Jaccard index of 0.6704$\pm$0.2167. These results illustrate that the proposed method shows promise for future work related to the segmentation and classification of benign and malignant breast lesions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge