Felix Meissen
NOVA: An Agentic Framework for Automated Histopathology Analysis and Discovery
Nov 14, 2025



Abstract:Digitized histopathology analysis involves complex, time-intensive workflows and specialized expertise, limiting its accessibility. We introduce NOVA, an agentic framework that translates scientific queries into executable analysis pipelines by iteratively generating and running Python code. NOVA integrates 49 domain-specific tools (e.g., nuclei segmentation, whole-slide encoding) built on open-source software, and can also create new tools ad hoc. To evaluate such systems, we present SlideQuest, a 90-question benchmark -- verified by pathologists and biomedical scientists -- spanning data processing, quantitative analysis, and hypothesis testing. Unlike prior biomedical benchmarks focused on knowledge recall or diagnostic QA, SlideQuest demands multi-step reasoning, iterative coding, and computational problem solving. Quantitative evaluation shows NOVA outperforms coding-agent baselines, and a pathologist-verified case study links morphology to prognostically relevant PAM50 subtypes, demonstrating its scalable discovery potential.
Evaluation of Language Models in the Medical Context Under Resource-Constrained Settings
Jun 24, 2024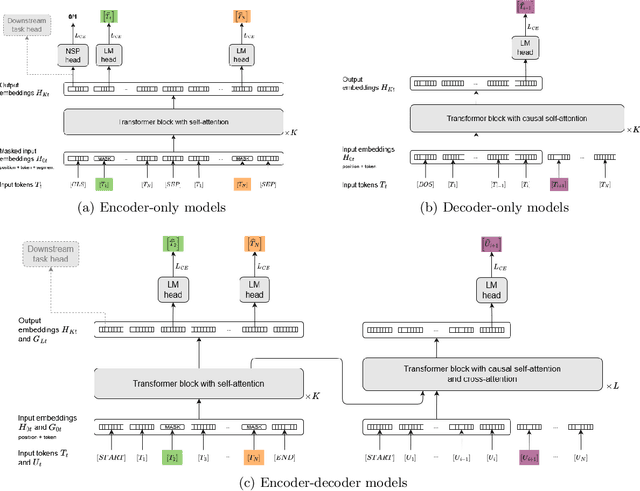
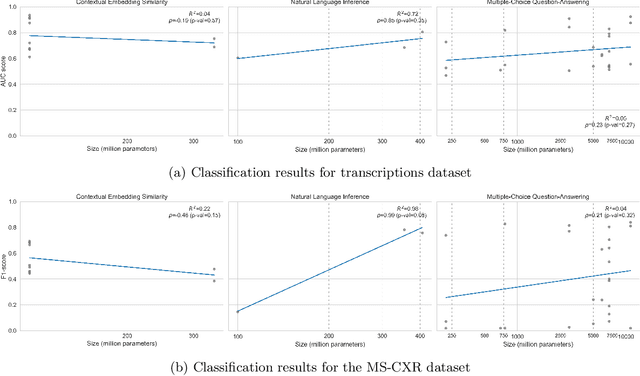
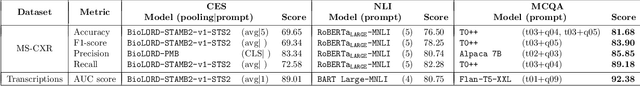
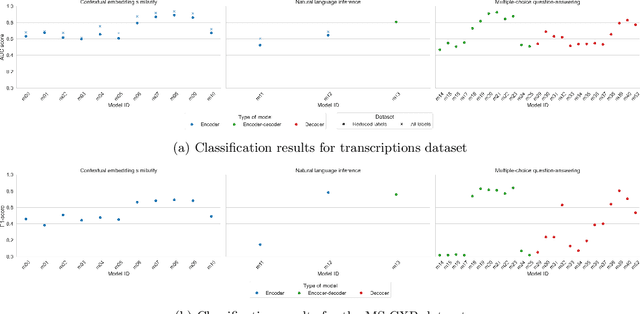
Abstract:Since the emergence of the Transformer architecture, language model development has increased, driven by their promising potential. However, releasing these models into production requires properly understanding their behavior, particularly in sensitive domains such as medicine. Despite this need, the medical literature still lacks technical assessments of pre-trained language models, which are especially valuable in resource-constrained settings in terms of computational power or limited budget. To address this gap, we provide a comprehensive survey of language models in the medical domain. In addition, we selected a subset of these models for thorough evaluation, focusing on classification and text generation tasks. Our subset encompasses 53 models, ranging from 110 million to 13 billion parameters, spanning the three families of Transformer-based models and from diverse knowledge domains. This study employs a series of approaches for text classification together with zero-shot prompting instead of model training or fine-tuning, which closely resembles the limited resource setting in which many users of language models find themselves. Encouragingly, our findings reveal remarkable performance across various tasks and datasets, underscoring the latent potential of certain models to contain medical knowledge, even without domain specialization. Consequently, our study advocates for further exploration of model applications in medical contexts, particularly in resource-constrained settings. The code is available on https://github.com/anpoc/Language-models-in-medicine.
MAIRA-2: Grounded Radiology Report Generation
Jun 06, 2024



Abstract:Radiology reporting is a complex task that requires detailed image understanding, integration of multiple inputs, including comparison with prior imaging, and precise language generation. This makes it ideal for the development and use of generative multimodal models. Here, we extend report generation to include the localisation of individual findings on the image - a task we call grounded report generation. Prior work indicates that grounding is important for clarifying image understanding and interpreting AI-generated text. Therefore, grounded reporting stands to improve the utility and transparency of automated report drafting. To enable evaluation of grounded reporting, we propose a novel evaluation framework - RadFact - leveraging the reasoning capabilities of large language models (LLMs). RadFact assesses the factuality of individual generated sentences, as well as correctness of generated spatial localisations when present. We introduce MAIRA-2, a large multimodal model combining a radiology-specific image encoder with a LLM, and trained for the new task of grounded report generation on chest X-rays. MAIRA-2 uses more comprehensive inputs than explored previously: the current frontal image, the current lateral image, the prior frontal image and prior report, as well as the Indication, Technique and Comparison sections of the current report. We demonstrate that these additions significantly improve report quality and reduce hallucinations, establishing a new state of the art on findings generation (without grounding) on MIMIC-CXR while demonstrating the feasibility of grounded reporting as a novel and richer task.
Weakly Supervised Object Detection in Chest X-Rays with Differentiable ROI Proposal Networks and Soft ROI Pooling
Feb 19, 2024



Abstract:Weakly supervised object detection (WSup-OD) increases the usefulness and interpretability of image classification algorithms without requiring additional supervision. The successes of multiple instance learning in this task for natural images, however, do not translate well to medical images due to the very different characteristics of their objects (i.e. pathologies). In this work, we propose Weakly Supervised ROI Proposal Networks (WSRPN), a new method for generating bounding box proposals on the fly using a specialized region of interest-attention (ROI-attention) module. WSRPN integrates well with classic backbone-head classification algorithms and is end-to-end trainable with only image-label supervision. We experimentally demonstrate that our new method outperforms existing methods in the challenging task of disease localization in chest X-ray images. Code: https://github.com/philip-mueller/wsrpn
How Low Can You Go? Surfacing Prototypical In-Distribution Samples for Unsupervised Anomaly Detection
Dec 06, 2023
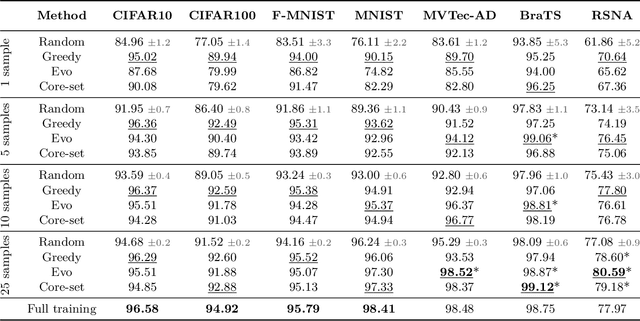
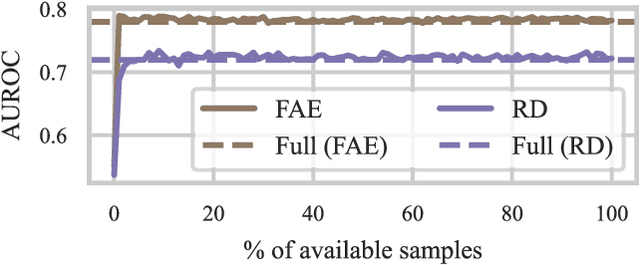
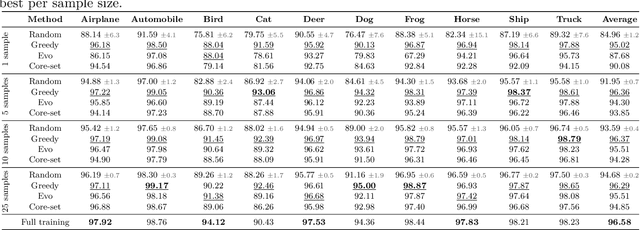
Abstract:Unsupervised anomaly detection (UAD) alleviates large labeling efforts by training exclusively on unlabeled in-distribution data and detecting outliers as anomalies. Generally, the assumption prevails that large training datasets allow the training of higher-performing UAD models. However, in this work, we show that using only very few training samples can already match - and in some cases even improve - anomaly detection compared to training with the whole training dataset. We propose three methods to identify prototypical samples from a large dataset of in-distribution samples. We demonstrate that by training with a subset of just ten such samples, we achieve an area under the receiver operating characteristics curve (AUROC) of $96.37 \%$ on CIFAR10, $92.59 \%$ on CIFAR100, $95.37 \%$ on MNIST, $95.38 \%$ on Fashion-MNIST, $96.37 \%$ on MVTec-AD, $98.81 \%$ on BraTS, and $81.95 \%$ on RSNA pneumonia detection, even exceeding the performance of full training in $25/67$ classes we tested. Additionally, we show that the prototypical in-distribution samples identified by our proposed methods translate well to different models and other datasets and that using their characteristics as guidance allows for successful manual selection of small subsets of high-performing samples. Our code is available at https://anonymous.4open.science/r/uad_prototypical_samples/
(Predictable) Performance Bias in Unsupervised Anomaly Detection
Sep 25, 2023



Abstract:Background: With the ever-increasing amount of medical imaging data, the demand for algorithms to assist clinicians has amplified. Unsupervised anomaly detection (UAD) models promise to aid in the crucial first step of disease detection. While previous studies have thoroughly explored fairness in supervised models in healthcare, for UAD, this has so far been unexplored. Methods: In this study, we evaluated how dataset composition regarding subgroups manifests in disparate performance of UAD models along multiple protected variables on three large-scale publicly available chest X-ray datasets. Our experiments were validated using two state-of-the-art UAD models for medical images. Finally, we introduced a novel subgroup-AUROC (sAUROC) metric, which aids in quantifying fairness in machine learning. Findings: Our experiments revealed empirical "fairness laws" (similar to "scaling laws" for Transformers) for training-dataset composition: Linear relationships between anomaly detection performance within a subpopulation and its representation in the training data. Our study further revealed performance disparities, even in the case of balanced training data, and compound effects that exacerbate the drop in performance for subjects associated with multiple adversely affected groups. Interpretation: Our study quantified the disparate performance of UAD models against certain demographic subgroups. Importantly, we showed that this unfairness cannot be mitigated by balanced representation alone. Instead, the representation of some subgroups seems harder to learn by UAD models than that of others. The empirical fairness laws discovered in our study make disparate performance in UAD models easier to estimate and aid in determining the most desirable dataset composition.
Anatomy-Driven Pathology Detection on Chest X-rays
Sep 05, 2023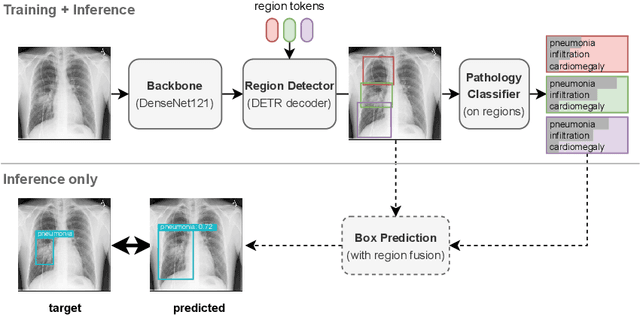
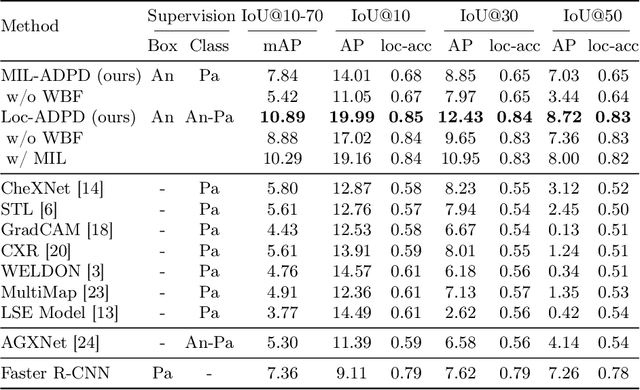

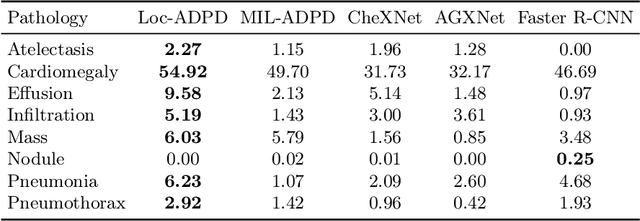
Abstract:Pathology detection and delineation enables the automatic interpretation of medical scans such as chest X-rays while providing a high level of explainability to support radiologists in making informed decisions. However, annotating pathology bounding boxes is a time-consuming task such that large public datasets for this purpose are scarce. Current approaches thus use weakly supervised object detection to learn the (rough) localization of pathologies from image-level annotations, which is however limited in performance due to the lack of bounding box supervision. We therefore propose anatomy-driven pathology detection (ADPD), which uses easy-to-annotate bounding boxes of anatomical regions as proxies for pathologies. We study two training approaches: supervised training using anatomy-level pathology labels and multiple instance learning (MIL) with image-level pathology labels. Our results show that our anatomy-level training approach outperforms weakly supervised methods and fully supervised detection with limited training samples, and our MIL approach is competitive with both baseline approaches, therefore demonstrating the potential of our approach.
The Brain Tumor Segmentation Challenge 2023: Brain MR Image Synthesis for Tumor Segmentation
May 20, 2023
Abstract:Automated brain tumor segmentation methods are well established, reaching performance levels with clear clinical utility. Most algorithms require four input magnetic resonance imaging (MRI) modalities, typically T1-weighted images with and without contrast enhancement, T2-weighted images, and FLAIR images. However, some of these sequences are often missing in clinical practice, e.g., because of time constraints and/or image artifacts (such as patient motion). Therefore, substituting missing modalities to recover segmentation performance in these scenarios is highly desirable and necessary for the more widespread adoption of such algorithms in clinical routine. In this work, we report the set-up of the Brain MR Image Synthesis Benchmark (BraSyn), organized in conjunction with the Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2023. The objective of the challenge is to benchmark image synthesis methods that realistically synthesize missing MRI modalities given multiple available images to facilitate automated brain tumor segmentation pipelines. The image dataset is multi-modal and diverse, created in collaboration with various hospitals and research institutions.
The Brain Tumor Segmentation Challenge 2023: Local Synthesis of Healthy Brain Tissue via Inpainting
May 15, 2023



Abstract:A myriad of algorithms for the automatic analysis of brain MR images is available to support clinicians in their decision-making. For brain tumor patients, the image acquisition time series typically starts with a scan that is already pathological. This poses problems, as many algorithms are designed to analyze healthy brains and provide no guarantees for images featuring lesions. Examples include but are not limited to algorithms for brain anatomy parcellation, tissue segmentation, and brain extraction. To solve this dilemma, we introduce the BraTS 2023 inpainting challenge. Here, the participants' task is to explore inpainting techniques to synthesize healthy brain scans from lesioned ones. The following manuscript contains the task formulation, dataset, and submission procedure. Later it will be updated to summarize the findings of the challenge. The challenge is organized as part of the BraTS 2023 challenge hosted at the MICCAI 2023 conference in Vancouver, Canada.
Robust Detection Outcome: A Metric for Pathology Detection in Medical Images
Mar 03, 2023Abstract:Detection of pathologies is a fundamental task in medical imaging and the evaluation of algorithms that can perform this task automatically is crucial. However, current object detection metrics for natural images do not reflect the specific clinical requirements in pathology detection sufficiently. To tackle this problem, we propose Robust Detection Outcome (RoDeO); a novel metric for evaluating algorithms for pathology detection in medical images, especially in chest X-rays. RoDeO evaluates different errors directly and individually, and reflects clinical needs better than current metrics. Extensive evaluation on the ChestX-ray8 dataset shows the superiority of our metrics compared to existing ones. We released the code at https://github.com/FeliMe/RoDeO and published RoDeO as pip package (rodeometric).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge