Felix Steinbauer
Grokking in the Wild: Data Augmentation for Real-World Multi-Hop Reasoning with Transformers
Apr 29, 2025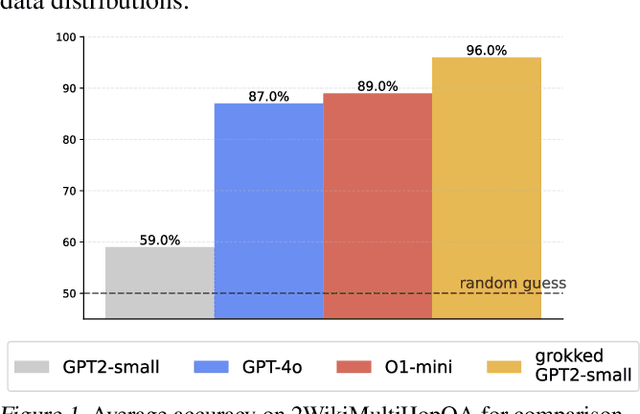

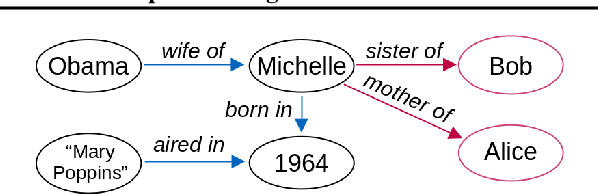

Abstract:Transformers have achieved great success in numerous NLP tasks but continue to exhibit notable gaps in multi-step factual reasoning, especially when real-world knowledge is sparse. Recent advances in grokking have demonstrated that neural networks can transition from memorizing to perfectly generalizing once they detect underlying logical patterns - yet these studies have primarily used small, synthetic tasks. In this paper, for the first time, we extend grokking to real-world factual data and address the challenge of dataset sparsity by augmenting existing knowledge graphs with carefully designed synthetic data to raise the ratio $\phi_r$ of inferred facts to atomic facts above the threshold required for grokking. Surprisingly, we find that even factually incorrect synthetic data can strengthen emergent reasoning circuits rather than degrade accuracy, as it forces the model to rely on relational structure rather than memorization. When evaluated on multi-hop reasoning benchmarks, our approach achieves up to 95-100% accuracy on 2WikiMultiHopQA - substantially improving over strong baselines and matching or exceeding current state-of-the-art results. We further provide an in-depth analysis of how increasing $\phi_r$ drives the formation of generalizing circuits inside Transformers. Our findings suggest that grokking-based data augmentation can unlock implicit multi-hop reasoning capabilities, opening the door to more robust and interpretable factual reasoning in large-scale language models.
P-TA: Using Proximal Policy Optimization to Enhance Tabular Data Augmentation via Large Language Models
Jun 17, 2024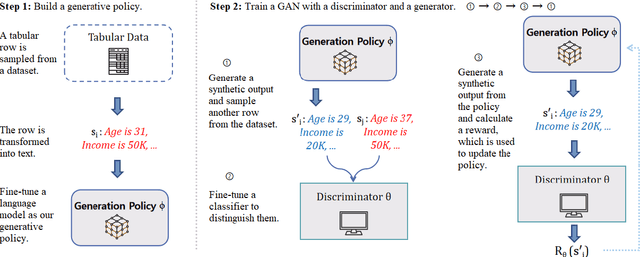



Abstract:A multitude of industries depend on accurate and reasonable tabular data augmentation for their business processes. Contemporary methodologies in generating tabular data revolve around utilizing Generative Adversarial Networks (GAN) or fine-tuning Large Language Models (LLM). However, GAN-based approaches are documented to produce samples with common-sense errors attributed to the absence of external knowledge. On the other hand, LLM-based methods exhibit a limited capacity to capture the disparities between synthesized and actual data distribution due to the absence of feedback from a discriminator during training. Furthermore, the decoding of LLM-based generation introduces gradient breakpoints, impeding the backpropagation of loss from a discriminator, thereby complicating the integration of these two approaches. To solve this challenge, we propose using proximal policy optimization (PPO) to apply GANs, guiding LLMs to enhance the probability distribution of tabular features. This approach enables the utilization of LLMs as generators for GANs in synthesizing tabular data. Our experiments demonstrate that PPO leads to an approximately 4\% improvement in the accuracy of models trained on synthetically generated data over state-of-the-art across three real-world datasets.
The Brain Tumor Segmentation Challenge 2023: Local Synthesis of Healthy Brain Tissue via Inpainting
May 15, 2023



Abstract:A myriad of algorithms for the automatic analysis of brain MR images is available to support clinicians in their decision-making. For brain tumor patients, the image acquisition time series typically starts with a scan that is already pathological. This poses problems, as many algorithms are designed to analyze healthy brains and provide no guarantees for images featuring lesions. Examples include but are not limited to algorithms for brain anatomy parcellation, tissue segmentation, and brain extraction. To solve this dilemma, we introduce the BraTS 2023 inpainting challenge. Here, the participants' task is to explore inpainting techniques to synthesize healthy brain scans from lesioned ones. The following manuscript contains the task formulation, dataset, and submission procedure. Later it will be updated to summarize the findings of the challenge. The challenge is organized as part of the BraTS 2023 challenge hosted at the MICCAI 2023 conference in Vancouver, Canada.
Learn-Morph-Infer: a new way of solving the inverse problem for brain tumor modeling
Nov 07, 2021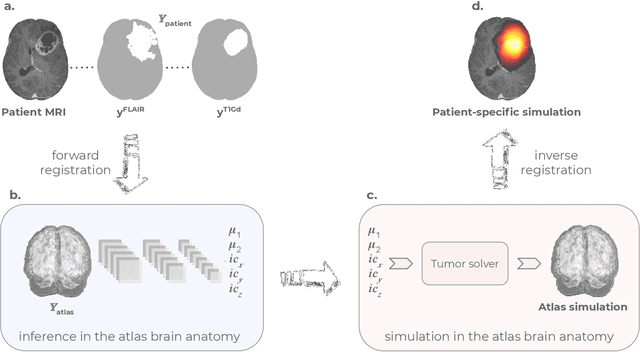
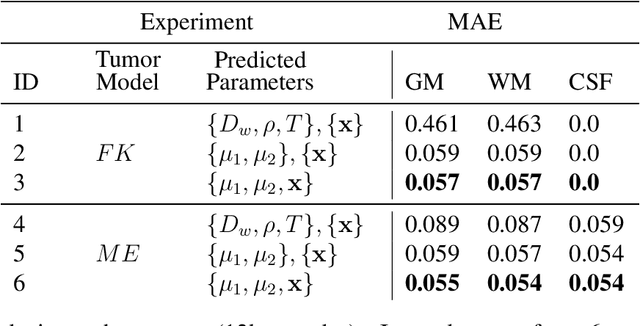
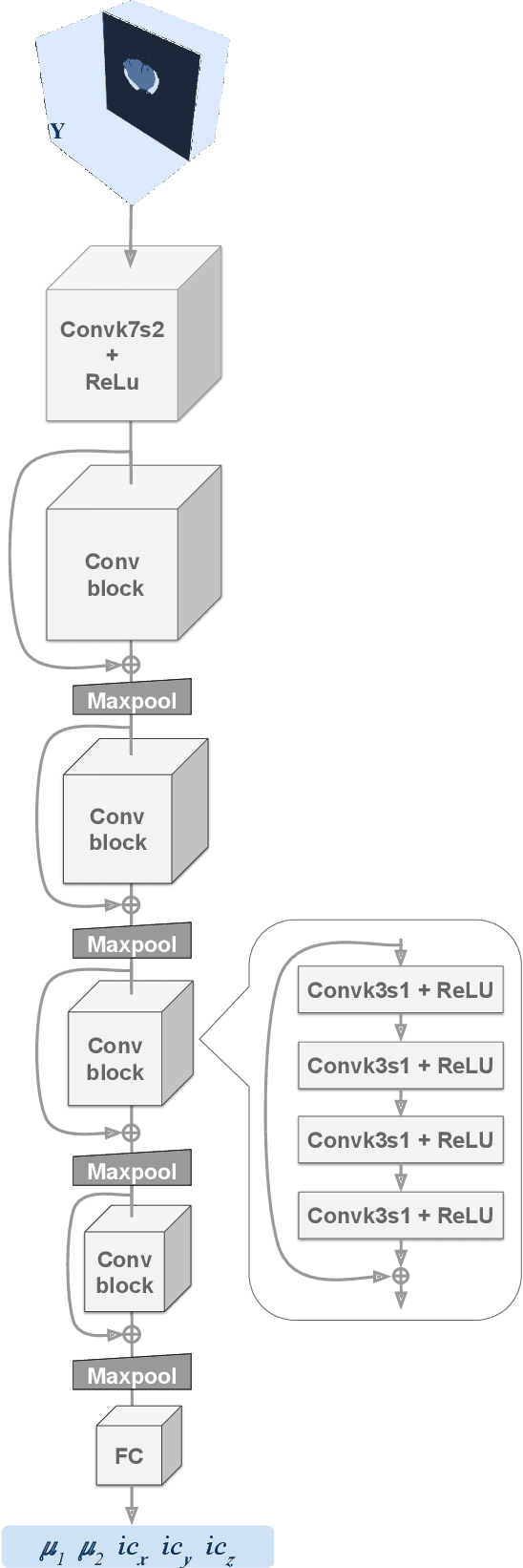
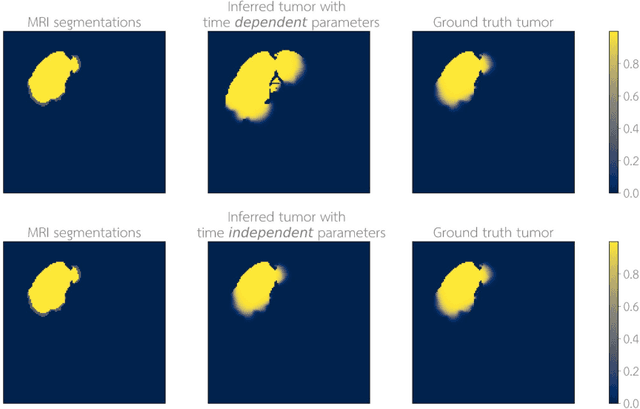
Abstract:Current treatment planning of patients diagnosed with brain tumor could significantly benefit by accessing the spatial distribution of tumor cell concentration. Existing diagnostic modalities, such as magnetic-resonance imaging (MRI), contrast sufficiently well areas of high cell density. However, they do not portray areas of low concentration, which can often serve as a source for the secondary appearance of the tumor after treatment. Numerical simulations of tumor growth could complement imaging information by providing estimates of full spatial distributions of tumor cells. Over recent years a corpus of literature on medical image-based tumor modeling was published. It includes different mathematical formalisms describing the forward tumor growth model. Alongside, various parametric inference schemes were developed to perform an efficient tumor model personalization, i.e. solving the inverse problem. However, the unifying drawback of all existing approaches is the time complexity of the model personalization that prohibits a potential integration of the modeling into clinical settings. In this work, we introduce a methodology for inferring patient-specific spatial distribution of brain tumor from T1Gd and FLAIR MRI medical scans. Coined as \textit{Learn-Morph-Infer} the method achieves real-time performance in the order of minutes on widely available hardware and the compute time is stable across tumor models of different complexity, such as reaction-diffusion and reaction-advection-diffusion models. We believe the proposed inverse solution approach not only bridges the way for clinical translation of brain tumor personalization but can also be adopted to other scientific and engineering domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge