Sam Bond-Taylor
Data Scaling Laws for Radiology Foundation Models
Sep 16, 2025Abstract:Foundation vision encoders such as CLIP and DINOv2, trained on web-scale data, exhibit strong transfer performance across tasks and datasets. However, medical imaging foundation models remain constrained by smaller datasets, limiting our understanding of how data scale and pretraining paradigms affect performance in this setting. In this work, we systematically study continual pretraining of two vision encoders, MedImageInsight (MI2) and RAD-DINO representing the two major encoder paradigms CLIP and DINOv2, on up to 3.5M chest x-rays from a single institution, holding compute and evaluation protocols constant. We evaluate on classification (radiology findings, lines and tubes), segmentation (lines and tubes), and radiology report generation. While prior work has primarily focused on tasks related to radiology findings, we include lines and tubes tasks to counterbalance this bias and evaluate a model's ability to extract features that preserve continuity along elongated structures. Our experiments show that MI2 scales more effectively for finding-related tasks, while RAD-DINO is stronger on tube-related tasks. Surprisingly, continually pretraining MI2 with both reports and structured labels using UniCL improves performance, underscoring the value of structured supervision at scale. We further show that for some tasks, as few as 30k in-domain samples are sufficient to surpass open-weights foundation models. These results highlight the utility of center-specific continual pretraining, enabling medical institutions to derive significant performance gains by utilizing in-domain data.
MAIRA-Seg: Enhancing Radiology Report Generation with Segmentation-Aware Multimodal Large Language Models
Nov 18, 2024



Abstract:There is growing interest in applying AI to radiology report generation, particularly for chest X-rays (CXRs). This paper investigates whether incorporating pixel-level information through segmentation masks can improve fine-grained image interpretation of multimodal large language models (MLLMs) for radiology report generation. We introduce MAIRA-Seg, a segmentation-aware MLLM framework designed to utilize semantic segmentation masks alongside CXRs for generating radiology reports. We train expert segmentation models to obtain mask pseudolabels for radiology-specific structures in CXRs. Subsequently, building on the architectures of MAIRA, a CXR-specialised model for report generation, we integrate a trainable segmentation tokens extractor that leverages these mask pseudolabels, and employ mask-aware prompting to generate draft radiology reports. Our experiments on the publicly available MIMIC-CXR dataset show that MAIRA-Seg outperforms non-segmentation baselines. We also investigate set-of-marks prompting with MAIRA and find that MAIRA-Seg consistently demonstrates comparable or superior performance. The results confirm that using segmentation masks enhances the nuanced reasoning of MLLMs, potentially contributing to better clinical outcomes.
MAIRA-2: Grounded Radiology Report Generation
Jun 06, 2024



Abstract:Radiology reporting is a complex task that requires detailed image understanding, integration of multiple inputs, including comparison with prior imaging, and precise language generation. This makes it ideal for the development and use of generative multimodal models. Here, we extend report generation to include the localisation of individual findings on the image - a task we call grounded report generation. Prior work indicates that grounding is important for clarifying image understanding and interpreting AI-generated text. Therefore, grounded reporting stands to improve the utility and transparency of automated report drafting. To enable evaluation of grounded reporting, we propose a novel evaluation framework - RadFact - leveraging the reasoning capabilities of large language models (LLMs). RadFact assesses the factuality of individual generated sentences, as well as correctness of generated spatial localisations when present. We introduce MAIRA-2, a large multimodal model combining a radiology-specific image encoder with a LLM, and trained for the new task of grounded report generation on chest X-rays. MAIRA-2 uses more comprehensive inputs than explored previously: the current frontal image, the current lateral image, the prior frontal image and prior report, as well as the Indication, Technique and Comparison sections of the current report. We demonstrate that these additions significantly improve report quality and reduce hallucinations, establishing a new state of the art on findings generation (without grounding) on MIMIC-CXR while demonstrating the feasibility of grounded reporting as a novel and richer task.
RAD-DINO: Exploring Scalable Medical Image Encoders Beyond Text Supervision
Jan 19, 2024Abstract:Language-supervised pre-training has proven to be a valuable method for extracting semantically meaningful features from images, serving as a foundational element in multimodal systems within the computer vision and medical imaging domains. However, resulting features are limited by the information contained within the text. This is particularly problematic in medical imaging, where radiologists' written findings focus on specific observations; a challenge compounded by the scarcity of paired imaging-text data due to concerns over leakage of personal health information. In this work, we fundamentally challenge the prevailing reliance on language supervision for learning general purpose biomedical imaging encoders. We introduce RAD-DINO, a biomedical image encoder pre-trained solely on unimodal biomedical imaging data that obtains similar or greater performance than state-of-the-art biomedical language supervised models on a diverse range of benchmarks. Specifically, the quality of learned representations is evaluated on standard imaging tasks (classification and semantic segmentation), and a vision-language alignment task (text report generation from images). To further demonstrate the drawback of language supervision, we show that features from RAD-DINO correlate with other medical records (e.g., sex or age) better than language-supervised models, which are generally not mentioned in radiology reports. Finally, we conduct a series of ablations determining the factors in RAD-DINO's performance; notably, we observe that RAD-DINO's downstream performance scales well with the quantity and diversity of training data, demonstrating that image-only supervision is a scalable approach for training a foundational biomedical image encoder.
RadEdit: stress-testing biomedical vision models via diffusion image editing
Dec 21, 2023Abstract:Biomedical imaging datasets are often small and biased, meaning that real-world performance of predictive models can be substantially lower than expected from internal testing. This work proposes using generative image editing to simulate dataset shifts and diagnose failure modes of biomedical vision models; this can be used in advance of deployment to assess readiness, potentially reducing cost and patient harm. Existing editing methods can produce undesirable changes, with spurious correlations learned due to the co-occurrence of disease and treatment interventions, limiting practical applicability. To address this, we train a text-to-image diffusion model on multiple chest X-ray datasets and introduce a new editing method RadEdit that uses multiple masks, if present, to constrain changes and ensure consistency in the edited images. We consider three types of dataset shifts: acquisition shift, manifestation shift, and population shift, and demonstrate that our approach can diagnose failures and quantify model robustness without additional data collection, complementing more qualitative tools for explainable AI.
Unaligned 2D to 3D Translation with Conditional Vector-Quantized Code Diffusion using Transformers
Aug 27, 2023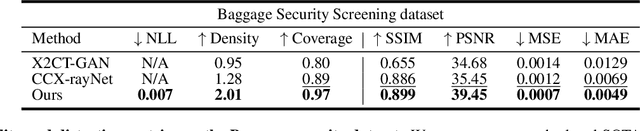
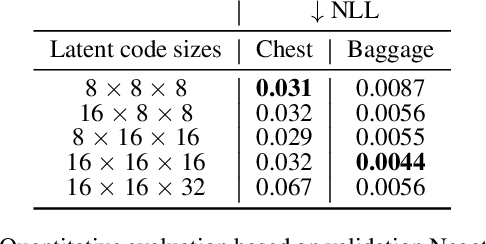
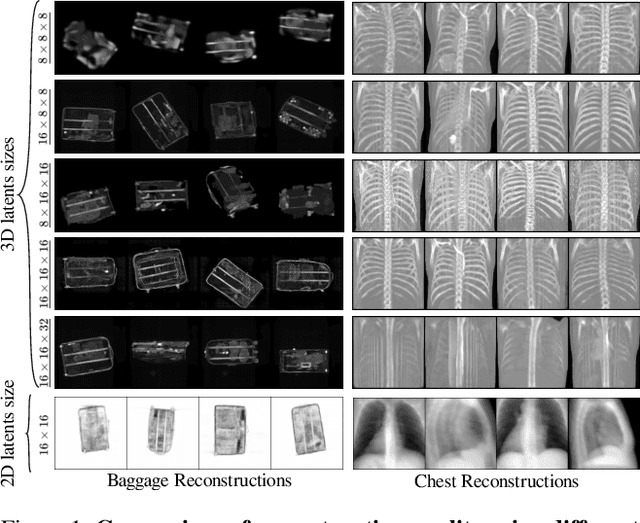
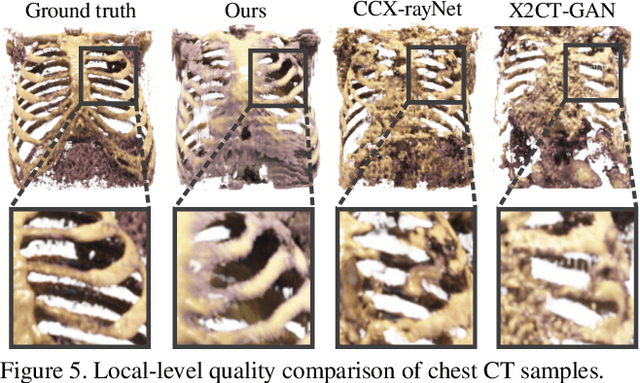
Abstract:Generating 3D images of complex objects conditionally from a few 2D views is a difficult synthesis problem, compounded by issues such as domain gap and geometric misalignment. For instance, a unified framework such as Generative Adversarial Networks cannot achieve this unless they explicitly define both a domain-invariant and geometric-invariant joint latent distribution, whereas Neural Radiance Fields are generally unable to handle both issues as they optimize at the pixel level. By contrast, we propose a simple and novel 2D to 3D synthesis approach based on conditional diffusion with vector-quantized codes. Operating in an information-rich code space enables high-resolution 3D synthesis via full-coverage attention across the views. Specifically, we generate the 3D codes (e.g. for CT images) conditional on previously generated 3D codes and the entire codebook of two 2D views (e.g. 2D X-rays). Qualitative and quantitative results demonstrate state-of-the-art performance over specialized methods across varied evaluation criteria, including fidelity metrics such as density, coverage, and distortion metrics for two complex volumetric imagery datasets from in real-world scenarios.
$\infty$-Diff: Infinite Resolution Diffusion with Subsampled Mollified States
Mar 31, 2023



Abstract:We introduce $\infty$-Diff, a generative diffusion model which directly operates on infinite resolution data. By randomly sampling subsets of coordinates during training and learning to denoise the content at those coordinates, a continuous function is learned that allows sampling at arbitrary resolutions. In contrast to other recent infinite resolution generative models, our approach operates directly on the raw data, not requiring latent vector compression for context, using hypernetworks, nor relying on discrete components. As such, our approach achieves significantly higher sample quality, as evidenced by lower FID scores, as well as being able to effectively scale to higher resolutions than the training data while retaining detail.
MedNeRF: Medical Neural Radiance Fields for Reconstructing 3D-aware CT-Projections from a Single X-ray
Feb 02, 2022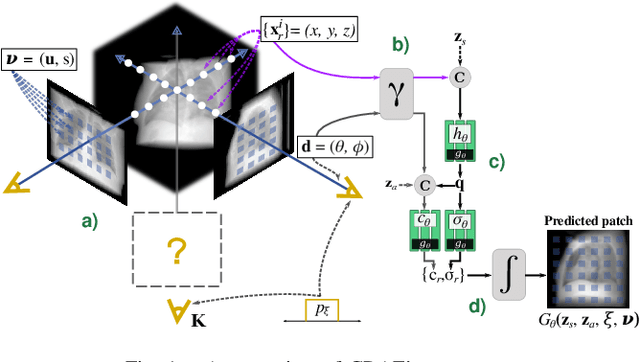
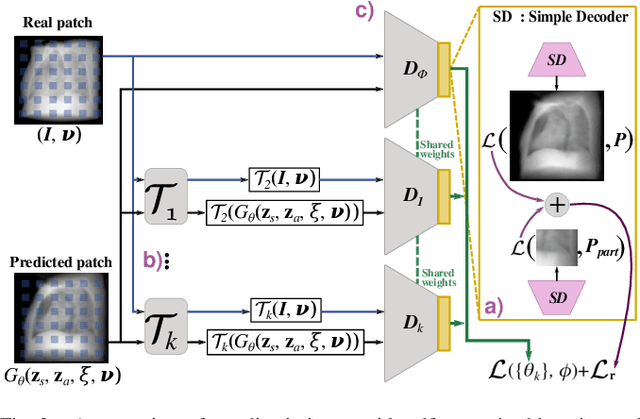
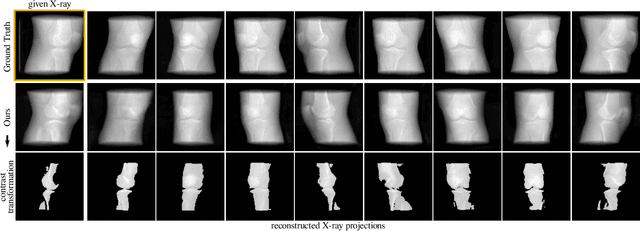
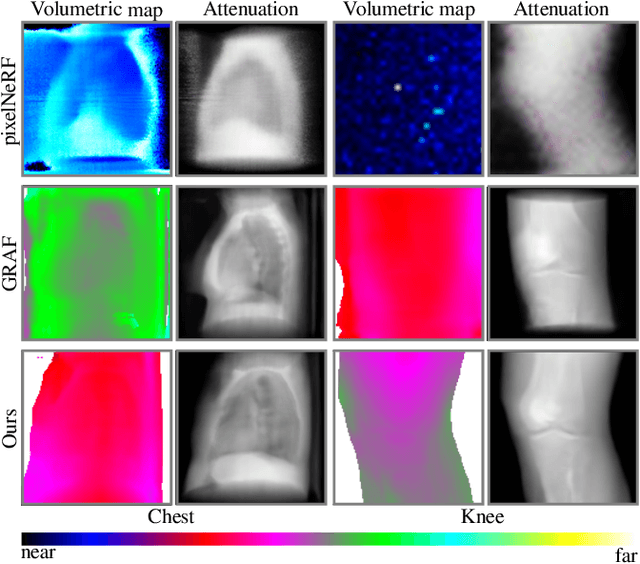
Abstract:Computed tomography (CT) is an effective medical imaging modality, widely used in the field of clinical medicine for the diagnosis of various pathologies. Advances in Multidetector CT imaging technology have enabled additional functionalities, including generation of thin slice multiplanar cross-sectional body imaging and 3D reconstructions. However, this involves patients being exposed to a considerable dose of ionising radiation. Excessive ionising radiation can lead to deterministic and harmful effects on the body. This paper proposes a Deep Learning model that learns to reconstruct CT projections from a few or even a single-view X-ray. This is based on a novel architecture that builds from neural radiance fields, which learns a continuous representation of CT scans by disentangling the shape and volumetric depth of surface and internal anatomical structures from 2D images. Our model is trained on chest and knee datasets, and we demonstrate qualitative and quantitative high-fidelity renderings and compare our approach to other recent radiance field-based methods. Our code and link to our datasets will be available at our GitHub.
Unleashing Transformers: Parallel Token Prediction with Discrete Absorbing Diffusion for Fast High-Resolution Image Generation from Vector-Quantized Codes
Nov 24, 2021

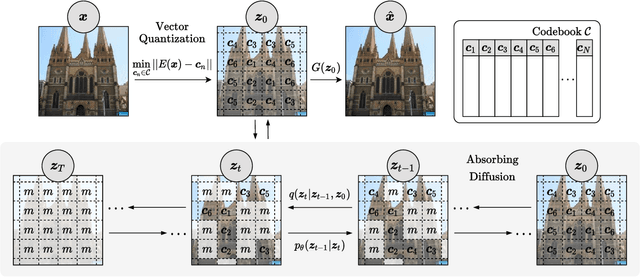

Abstract:Whilst diffusion probabilistic models can generate high quality image content, key limitations remain in terms of both generating high-resolution imagery and their associated high computational requirements. Recent Vector-Quantized image models have overcome this limitation of image resolution but are prohibitively slow and unidirectional as they generate tokens via element-wise autoregressive sampling from the prior. By contrast, in this paper we propose a novel discrete diffusion probabilistic model prior which enables parallel prediction of Vector-Quantized tokens by using an unconstrained Transformer architecture as the backbone. During training, tokens are randomly masked in an order-agnostic manner and the Transformer learns to predict the original tokens. This parallelism of Vector-Quantized token prediction in turn facilitates unconditional generation of globally consistent high-resolution and diverse imagery at a fraction of the computational expense. In this manner, we can generate image resolutions exceeding that of the original training set samples whilst additionally provisioning per-image likelihood estimates (in a departure from generative adversarial approaches). Our approach achieves state-of-the-art results in terms of Density (LSUN Bedroom: 1.51; LSUN Churches: 1.12; FFHQ: 1.20) and Coverage (LSUN Bedroom: 0.83; LSUN Churches: 0.73; FFHQ: 0.80), and performs competitively on FID (LSUN Bedroom: 3.64; LSUN Churches: 4.07; FFHQ: 6.11) whilst offering advantages in terms of both computation and reduced training set requirements.
Deep Generative Modelling: A Comparative Review of VAEs, GANs, Normalizing Flows, Energy-Based and Autoregressive Models
Mar 08, 2021



Abstract:Deep generative modelling is a class of techniques that train deep neural networks to model the distribution of training samples. Research has fragmented into various interconnected approaches, each of which making trade-offs including run-time, diversity, and architectural restrictions. In particular, this compendium covers energy-based models, variational autoencoders, generative adversarial networks, autoregressive models, normalizing flows, in addition to numerous hybrid approaches. These techniques are drawn under a single cohesive framework, comparing and contrasting to explain the premises behind each, while reviewing current state-of-the-art advances and implementations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge