Jonathan Frawley
MedNeRF: Medical Neural Radiance Fields for Reconstructing 3D-aware CT-Projections from a Single X-ray
Feb 02, 2022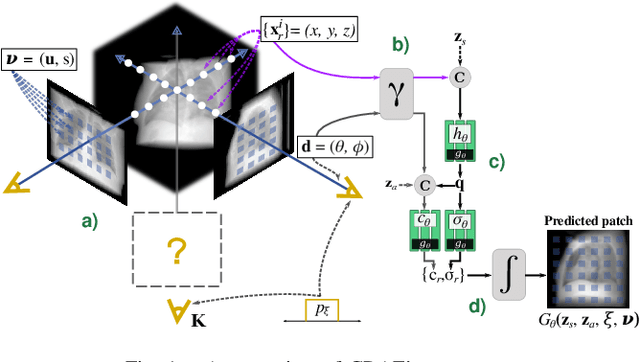
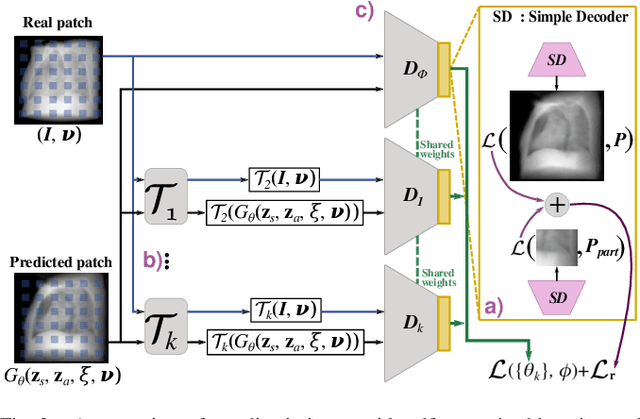
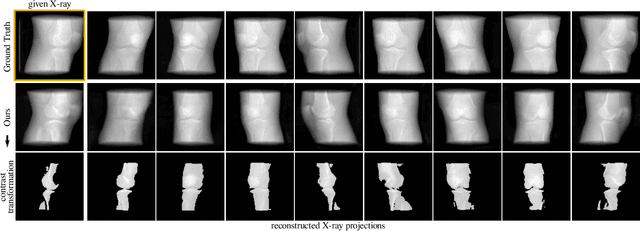
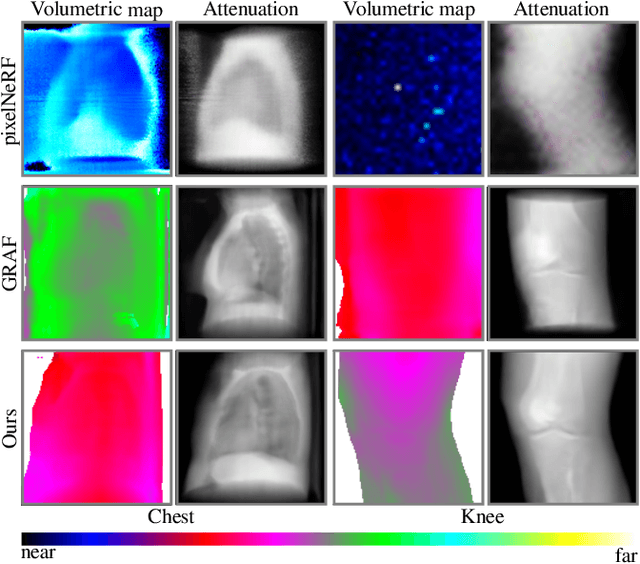
Abstract:Computed tomography (CT) is an effective medical imaging modality, widely used in the field of clinical medicine for the diagnosis of various pathologies. Advances in Multidetector CT imaging technology have enabled additional functionalities, including generation of thin slice multiplanar cross-sectional body imaging and 3D reconstructions. However, this involves patients being exposed to a considerable dose of ionising radiation. Excessive ionising radiation can lead to deterministic and harmful effects on the body. This paper proposes a Deep Learning model that learns to reconstruct CT projections from a few or even a single-view X-ray. This is based on a novel architecture that builds from neural radiance fields, which learns a continuous representation of CT scans by disentangling the shape and volumetric depth of surface and internal anatomical structures from 2D images. Our model is trained on chest and knee datasets, and we demonstrate qualitative and quantitative high-fidelity renderings and compare our approach to other recent radiance field-based methods. Our code and link to our datasets will be available at our GitHub.
Robust 3D U-Net Segmentation of Macular Holes
Mar 01, 2021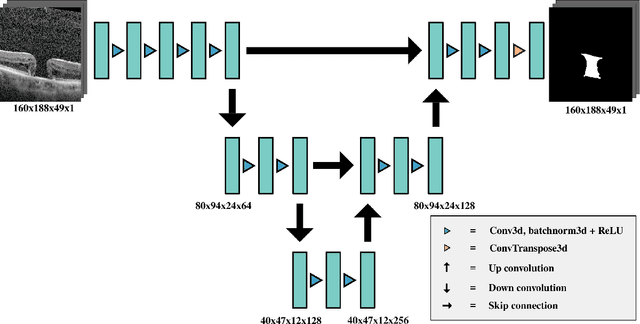
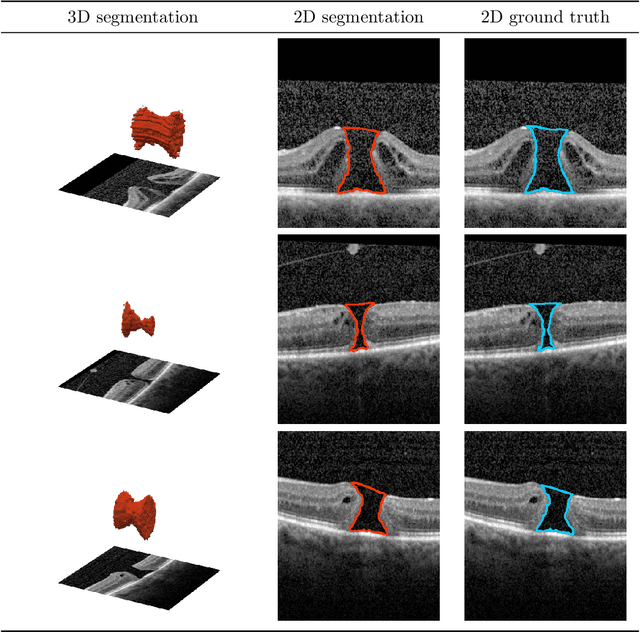
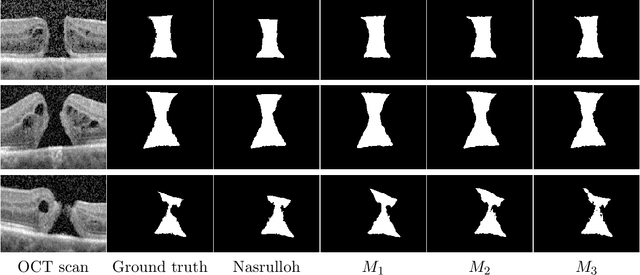
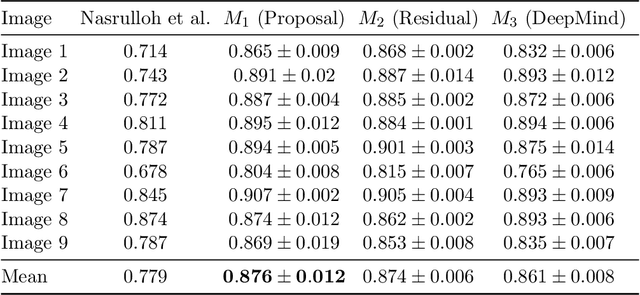
Abstract:Macular holes are a common eye condition which result in visual impairment. We look at the application of deep convolutional neural networks to the problem of macular hole segmentation. We use the 3D U-Net architecture as a basis and experiment with a number of design variants. Manually annotating and measuring macular holes is time consuming and error prone. Previous automated approaches to macular hole segmentation take minutes to segment a single 3D scan. Our proposed model generates significantly more accurate segmentations in less than a second. We found that an approach of architectural simplification, by greatly simplifying the network capacity and depth, exceeds both expert performance and state-of-the-art models such as residual 3D U-Nets.
Segmentation of Macular Edema Datasets with Small Residual 3D U-Net Architectures
May 10, 2020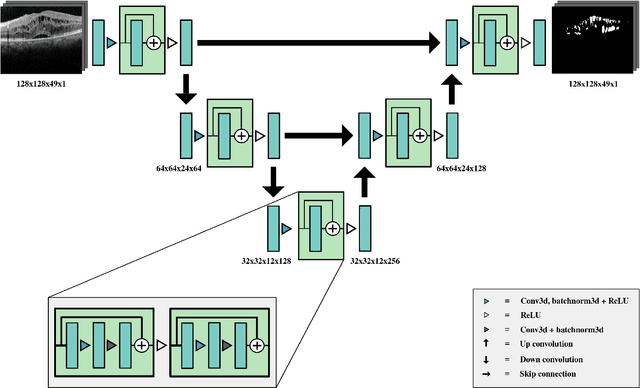
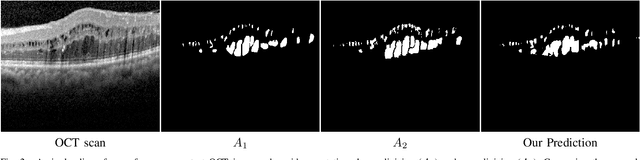
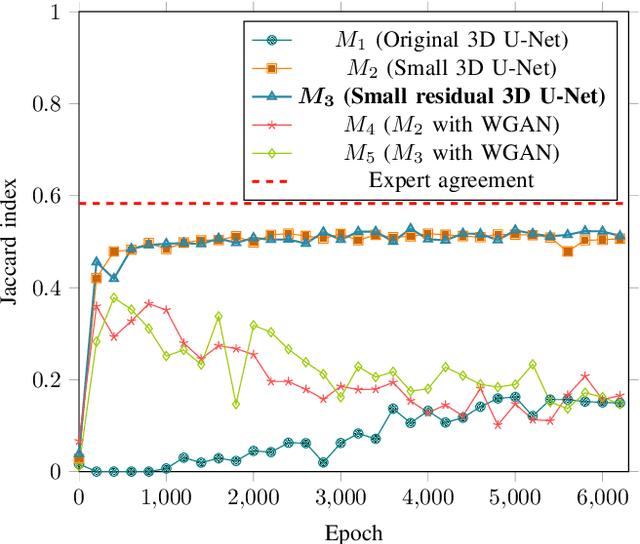
Abstract:This paper investigates the application of deep convolutional neural networks with prohibitively small datasets to the problem of macular edema segmentation. In particular, we investigate several different heavily regularized architectures. We find that, contrary to popular belief, neural architectures within this application setting are able to achieve close to human-level performance on unseen test images without requiring large numbers of training examples. Annotating these 3D datasets is difficult, with multiple criteria required. It takes an experienced clinician two days to annotate a single 3D image, whereas our trained model achieves similar performance in less than a second. We found that an approach which uses targeted dataset augmentation, alongside architectural simplification with an emphasis on residual design, has acceptable generalization performance - despite relying on fewer than 15 training examples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge