Johannes Brandt
Reconciling AI Performance and Data Reconstruction Resilience for Medical Imaging
Dec 05, 2023


Abstract:Artificial Intelligence (AI) models are vulnerable to information leakage of their training data, which can be highly sensitive, for example in medical imaging. Privacy Enhancing Technologies (PETs), such as Differential Privacy (DP), aim to circumvent these susceptibilities. DP is the strongest possible protection for training models while bounding the risks of inferring the inclusion of training samples or reconstructing the original data. DP achieves this by setting a quantifiable privacy budget. Although a lower budget decreases the risk of information leakage, it typically also reduces the performance of such models. This imposes a trade-off between robust performance and stringent privacy. Additionally, the interpretation of a privacy budget remains abstract and challenging to contextualize. In this study, we contrast the performance of AI models at various privacy budgets against both, theoretical risk bounds and empirical success of reconstruction attacks. We show that using very large privacy budgets can render reconstruction attacks impossible, while drops in performance are negligible. We thus conclude that not using DP -- at all -- is negligent when applying AI models to sensitive data. We deem those results to lie a foundation for further debates on striking a balance between privacy risks and model performance.
Anatomy-Driven Pathology Detection on Chest X-rays
Sep 05, 2023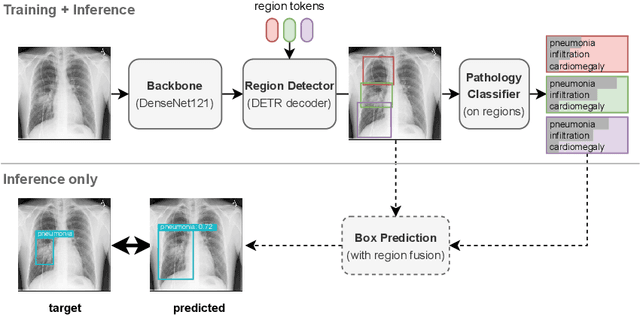
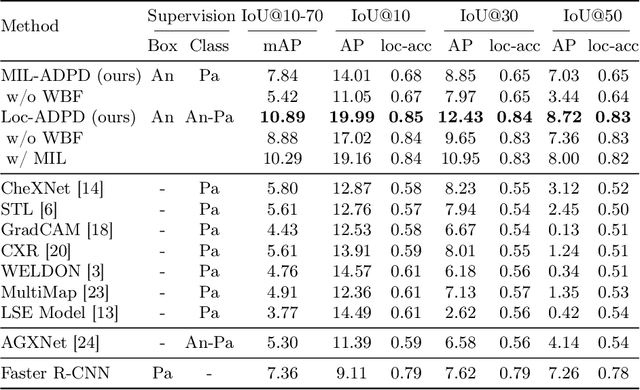

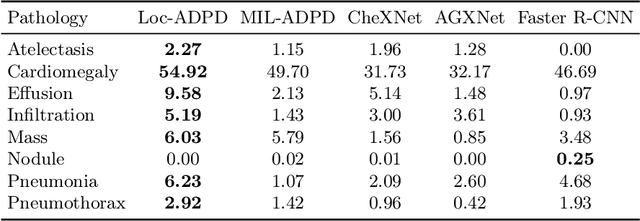
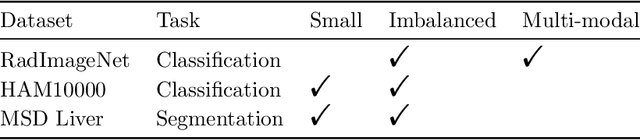
Abstract:Pathology detection and delineation enables the automatic interpretation of medical scans such as chest X-rays while providing a high level of explainability to support radiologists in making informed decisions. However, annotating pathology bounding boxes is a time-consuming task such that large public datasets for this purpose are scarce. Current approaches thus use weakly supervised object detection to learn the (rough) localization of pathologies from image-level annotations, which is however limited in performance due to the lack of bounding box supervision. We therefore propose anatomy-driven pathology detection (ADPD), which uses easy-to-annotate bounding boxes of anatomical regions as proxies for pathologies. We study two training approaches: supervised training using anatomy-level pathology labels and multiple instance learning (MIL) with image-level pathology labels. Our results show that our anatomy-level training approach outperforms weakly supervised methods and fully supervised detection with limited training samples, and our MIL approach is competitive with both baseline approaches, therefore demonstrating the potential of our approach.
Explainable 2D Vision Models for 3D Medical Data
Jul 13, 2023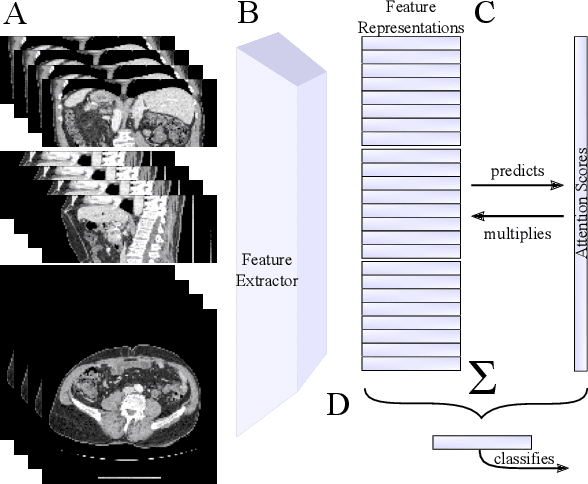
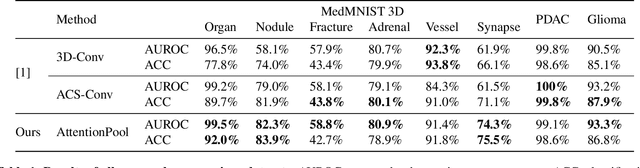
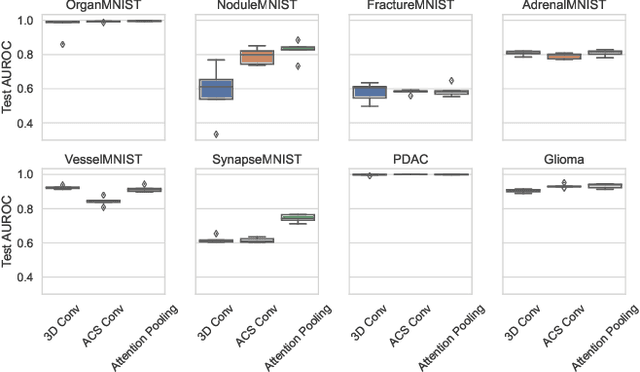
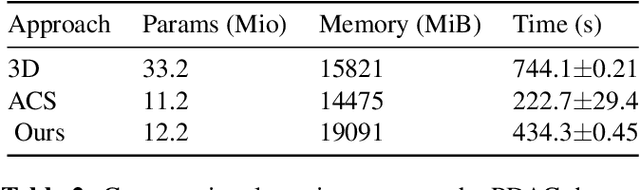
Abstract:Training Artificial Intelligence (AI) models on three-dimensional image data presents unique challenges compared to the two-dimensional case: Firstly, the computational resources are significantly higher, and secondly, the availability of large pretraining datasets is often limited, impeding training success. In this study, we propose a simple approach of adapting 2D networks with an intermediate feature representation for processing 3D volumes. Our method involves sequentially applying these networks to slices of a 3D volume from all orientations. Subsequently, a feature reduction module combines the extracted slice features into a single representation, which is then used for classification. We evaluate our approach on medical classification benchmarks and a real-world clinical dataset, demonstrating comparable results to existing methods. Furthermore, by employing attention pooling as a feature reduction module we obtain weighted importance values for each slice during the forward pass. We show that slices deemed important by our approach allow the inspection of the basis of a model's prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge