Harrison C. Gottlich
Role of Image Acquisition and Patient Phenotype Variations in Automatic Segmentation Model Generalization
Jul 26, 2023



Abstract:Purpose: This study evaluated the out-of-domain performance and generalization capabilities of automated medical image segmentation models, with a particular focus on adaptation to new image acquisitions and disease type. Materials: Datasets from both non-contrast and contrast-enhanced abdominal CT scans of healthy patients and those with polycystic kidney disease (PKD) were used. A total of 400 images (100 non-contrast controls, 100 contrast controls, 100 non-contrast PKD, 100 contrast PKD) were utilized for training/validation of models to segment kidneys, livers, and spleens, and the final models were then tested on 100 non-contrast CT images of patients affected by PKD. Performance was evaluated using Dice, Jaccard, TPR, and Precision. Results: Models trained on a diverse range of data showed no worse performance than models trained exclusively on in-domain data when tested on in-domain data. For instance, the Dice similarity of the model trained on 25% from each dataset was found to be non-inferior to the model trained purely on in-domain data. Conclusions: The results indicate that broader training examples significantly enhances model generalization and out-of-domain performance, thereby improving automated segmentation tools' applicability in clinical settings. The study's findings provide a roadmap for future research to adopt a data-centric approach in medical image AI model development.
AI in the Loop -- Functionalizing Fold Performance Disagreement to Monitor Automated Medical Image Segmentation Pipelines
May 15, 2023

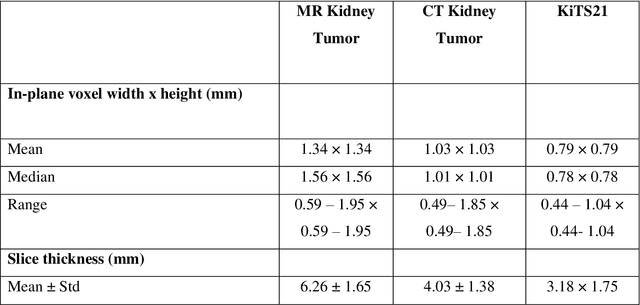

Abstract:Methods for automatically flag poor performing-predictions are essential for safely implementing machine learning workflows into clinical practice and for identifying difficult cases during model training. We present a readily adoptable method using sub-models trained on different dataset folds, where their disagreement serves as a surrogate for model confidence. Thresholds informed by human interobserver values were used to determine whether a final ensemble model prediction would require manual review. In two different datasets (abdominal CT and MR predicting kidney tumors), our framework effectively identified low performing automated segmentations. Flagging images with a minimum Interfold test Dice score below human interobserver variability maximized the number of flagged images while ensuring maximum ensemble test Dice. When our internally trained model was applied to an external publicly available dataset (KiTS21), flagged images included smaller tumors than those observed in our internally trained dataset, demonstrating the methods robustness to flagging poor performing out-of-distribution input data. Comparing interfold sub-model disagreement against human interobserver values is an efficient way to approximate a model's epistemic uncertainty - its lack of knowledge due to insufficient relevant training data - a key functionality for adopting these applications in clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge