Hierarchical Spatio-Temporal State-Space Modeling for fMRI Analysis
Paper and Code
Aug 23, 2024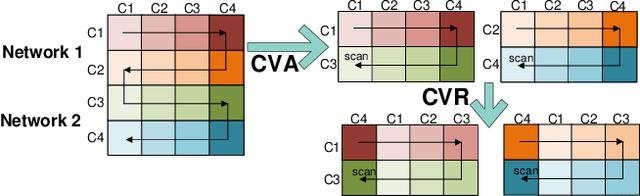
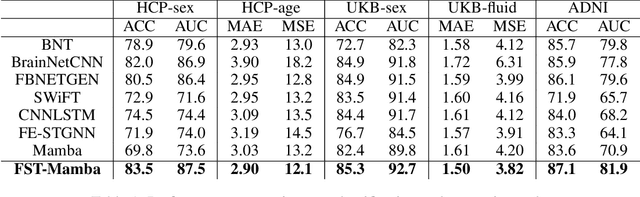
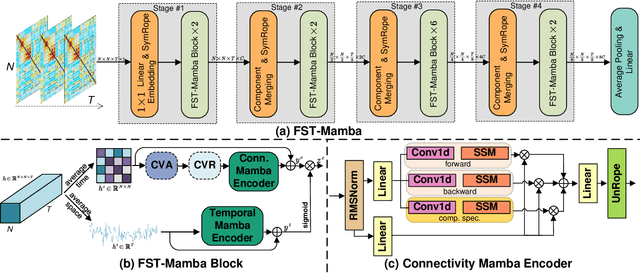

Recent advances in deep learning structured state space models, especially the Mamba architecture, have demonstrated remarkable performance improvements while maintaining linear complexity. In this study, we introduce functional spatiotemporal Mamba (FST-Mamba), a Mamba-based model designed for discovering neurological biomarkers using functional magnetic resonance imaging (fMRI). We focus on dynamic functional network connectivity (dFNC) derived from fMRI and propose a hierarchical spatiotemporal Mamba-based network that processes spatial and temporal information separately using Mamba-based encoders. Leveraging the topological uniqueness of the FNC matrix, we introduce a component-wise varied-scale aggregation (CVA) mechanism to aggregate connectivity across individual components within brain networks, enabling the model to capture both inter-component and inter-network information. To better handle the FNC data, we develop a new component-specific scanning order. Additionally, we propose symmetric rotary position encoding (SymRope) to encode the relative positions of each functional connection while considering the symmetric nature of the FNC matrix. Experimental results demonstrate significant improvements in the proposed FST-Mamba model on various brain-based classification and regression tasks. Our work reveals the substantial potential of attention-free sequence modeling in brain discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge