Zening Fu
Tri-Institutional Center for Translational Research in Neuroimaging and Data Science
An interpretable generative multimodal neuroimaging-genomics framework for decoding Alzheimer's disease
Jun 19, 2024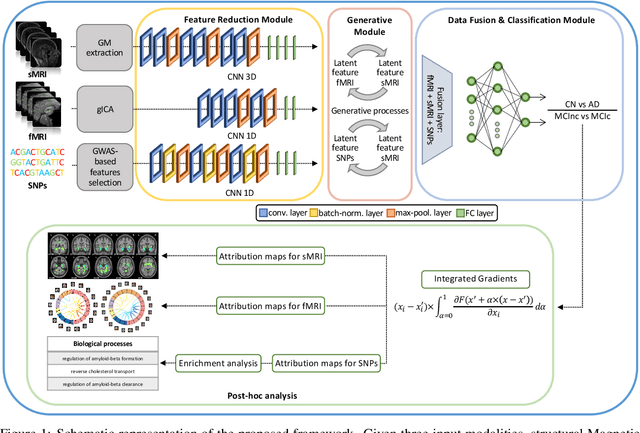

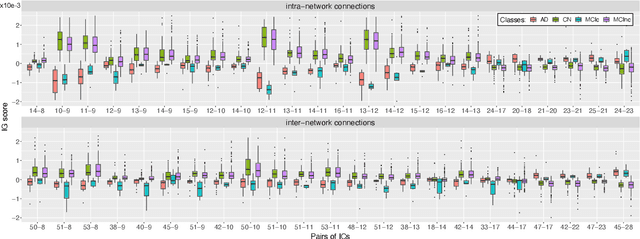

Abstract:Alzheimer's disease (AD) is the most prevalent form of dementia with a progressive decline in cognitive abilities. The AD continuum encompasses a prodormal stage known as Mild Cognitive Impairment (MCI), where patients may either progress to AD or remain stable. In this study, we leveraged structural and functional MRI to investigate the disease-induced grey matter and functional network connectivity changes. Moreover, considering AD's strong genetic component, we introduce SNPs as a third channel. Given such diverse inputs, missing one or more modalities is a typical concern of multimodal methods. We hence propose a novel deep learning-based classification framework where generative module employing Cycle GANs was adopted to impute missing data within the latent space. Additionally, we adopted an Explainable AI method, Integrated Gradients, to extract input features relevance, enhancing our understanding of the learned representations. Two critical tasks were addressed: AD detection and MCI conversion prediction. Experimental results showed that our model was able to reach the SOA in the classification of CN/AD reaching an average test accuracy of $0.926\pm0.02$. For the MCI task, we achieved an average prediction accuracy of $0.711\pm0.01$ using the pre-trained model for CN/AD. The interpretability analysis revealed significant grey matter modulations in cortical and subcortical brain areas well known for their association with AD. Moreover, impairments in sensory-motor and visual resting state network connectivity along the disease continuum, as well as mutations in SNPs defining biological processes linked to amyloid-beta and cholesterol formation clearance and regulation, were identified as contributors to the achieved performance. Overall, our integrative deep learning approach shows promise for AD detection and MCI prediction, while shading light on important biological insights.
An Interpretable Cross-Attentive Multi-modal MRI Fusion Framework for Schizophrenia Diagnosis
Mar 29, 2024



Abstract:Both functional and structural magnetic resonance imaging (fMRI and sMRI) are widely used for the diagnosis of mental disorder. However, combining complementary information from these two modalities is challenging due to their heterogeneity. Many existing methods fall short of capturing the interaction between these modalities, frequently defaulting to a simple combination of latent features. In this paper, we propose a novel Cross-Attentive Multi-modal Fusion framework (CAMF), which aims to capture both intra-modal and inter-modal relationships between fMRI and sMRI, enhancing multi-modal data representation. Specifically, our CAMF framework employs self-attention modules to identify interactions within each modality while cross-attention modules identify interactions between modalities. Subsequently, our approach optimizes the integration of latent features from both modalities. This approach significantly improves classification accuracy, as demonstrated by our evaluations on two extensive multi-modal brain imaging datasets, where CAMF consistently outperforms existing methods. Furthermore, the gradient-guided Score-CAM is applied to interpret critical functional networks and brain regions involved in schizophrenia. The bio-markers identified by CAMF align with established research, potentially offering new insights into the diagnosis and pathological endophenotypes of schizophrenia.
Multiscale Neuroimaging Features for the Identification of Medication Class and Non-Responders in Mood Disorder Treatment
Feb 12, 2024Abstract:In the clinical treatment of mood disorders, the complex behavioral symptoms presented by patients and variability of patient response to particular medication classes can create difficulties in providing fast and reliable treatment when standard diagnostic and prescription methods are used. Increasingly, the incorporation of physiological information such as neuroimaging scans and derivatives into the clinical process promises to alleviate some of the uncertainty surrounding this process. Particularly, if neural features can help to identify patients who may not respond to standard courses of anti-depressants or mood stabilizers, clinicians may elect to avoid lengthy and side-effect-laden treatments and seek out a different, more effective course that might otherwise not have been under consideration. Previously, approaches for the derivation of relevant neuroimaging features work at only one scale in the data, potentially limiting the depth of information available for clinical decision support. In this work, we show that the utilization of multi spatial scale neuroimaging features - particularly resting state functional networks and functional network connectivity measures - provide a rich and robust basis for the identification of relevant medication class and non-responders in the treatment of mood disorders. We demonstrate that the generated features, along with a novel approach for fast and automated feature selection, can support high accuracy rates in the identification of medication class and non-responders as well as the identification of novel, multi-scale biomarkers.
Self-Supervised Mental Disorder Classifiers via Time Reversal
Nov 30, 2022Abstract:Data scarcity is a notable problem, especially in the medical domain, due to patient data laws. Therefore, efficient Pre-Training techniques could help in combating this problem. In this paper, we demonstrate that a model trained on the time direction of functional neuro-imaging data could help in any downstream task, for example, classifying diseases from healthy controls in fMRI data. We train a Deep Neural Network on Independent components derived from fMRI data using the Independent component analysis (ICA) technique. It learns time direction in the ICA-based data. This pre-trained model is further trained to classify brain disorders in different datasets. Through various experiments, we have shown that learning time direction helps a model learn some causal relation in fMRI data that helps in faster convergence, and consequently, the model generalizes well in downstream classification tasks even with fewer data records.
MultiCrossViT: Multimodal Vision Transformer for Schizophrenia Prediction using Structural MRI and Functional Network Connectivity Data
Nov 12, 2022Abstract:Vision Transformer (ViT) is a pioneering deep learning framework that can address real-world computer vision issues, such as image classification and object recognition. Importantly, ViTs are proven to outperform traditional deep learning models, such as convolutional neural networks (CNNs). Relatively recently, a number of ViT mutations have been transplanted into the field of medical imaging, thereby resolving a variety of critical classification and segmentation challenges, especially in terms of brain imaging data. In this work, we provide a novel multimodal deep learning pipeline, MultiCrossViT, which is capable of analyzing both structural MRI (sMRI) and static functional network connectivity (sFNC) data for the prediction of schizophrenia disease. On a dataset with minimal training subjects, our novel model can achieve an AUC of 0.832. Finally, we visualize multiple brain regions and covariance patterns most relevant to schizophrenia based on the resulting ViT attention maps by extracting features from transformer encoders.
Prediction of Gender from Longitudinal MRI data via Deep Learning on Adolescent Data Reveals Unique Patterns Associated with Brain Structure and Change over a Two-year Period
Sep 15, 2022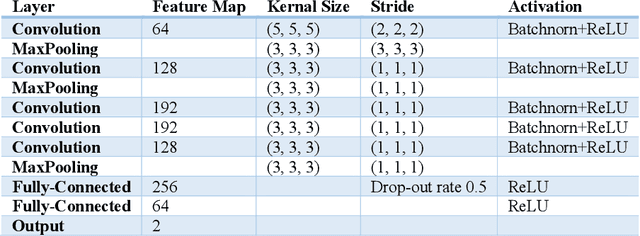
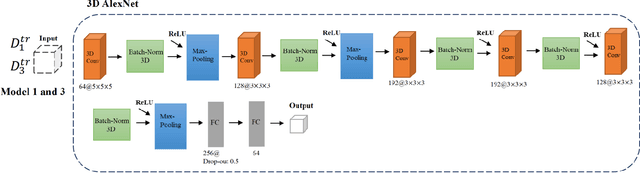
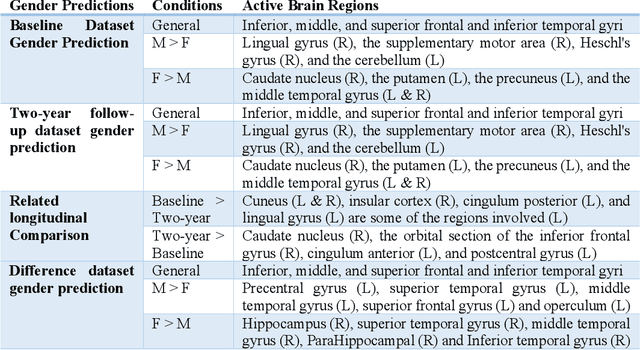
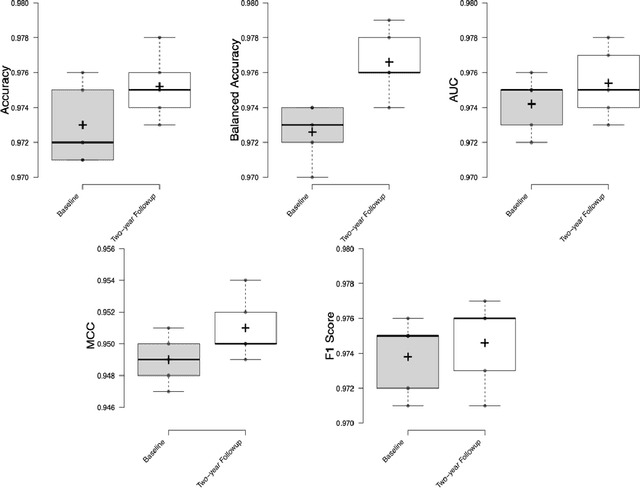
Abstract:Deep learning algorithms for predicting neuroimaging data have shown considerable promise in various applications. Prior work has demonstrated that deep learning models that take advantage of the data's 3D structure can outperform standard machine learning on several learning tasks. However, most prior research in this area has focused on neuroimaging data from adults. Within the Adolescent Brain and Cognitive Development (ABCD) dataset, a large longitudinal development study, we examine structural MRI data to predict gender and identify gender-related changes in brain structure. Results demonstrate that gender prediction accuracy is exceptionally high (>97%) with training epochs >200 and that this accuracy increases with age. Brain regions identified as the most discriminative in the task under study include predominantly frontal areas and the temporal lobe. When evaluating gender predictive changes specific to a two-year increase in age, a broader set of visual, cingulate, and insular regions are revealed. Our findings show a robust gender-related structural brain change pattern, even over a small age range. This suggests that it might be possible to study how the brain changes during adolescence by looking at how these changes are related to different behavioral and environmental factors.
Deep Dynamic Effective Connectivity Estimation from Multivariate Time Series
Feb 16, 2022



Abstract:Recently, methods that represent data as a graph, such as graph neural networks (GNNs) have been successfully used to learn data representations and structures to solve classification and link prediction problems. The applications of such methods are vast and diverse, but most of the current work relies on the assumption of a static graph. This assumption does not hold for many highly dynamic systems, where the underlying connectivity structure is non-stationary and is mostly unobserved. Using a static model in these situations may result in sub-optimal performance. In contrast, modeling changes in graph structure with time can provide information about the system whose applications go beyond classification. Most work of this type does not learn effective connectivity and focuses on cross-correlation between nodes to generate undirected graphs. An undirected graph is unable to capture direction of an interaction which is vital in many fields, including neuroscience. To bridge this gap, we developed dynamic effective connectivity estimation via neural network training (DECENNT), a novel model to learn an interpretable directed and dynamic graph induced by the downstream classification/prediction task. DECENNT outperforms state-of-the-art (SOTA) methods on five different tasks and infers interpretable task-specific dynamic graphs. The dynamic graphs inferred from functional neuroimaging data align well with the existing literature and provide additional information. Additionally, the temporal attention module of DECENNT identifies time-intervals crucial for predictive downstream task from multivariate time series data.
A deep learning model for data-driven discovery of functional connectivity
Dec 07, 2021



Abstract:Functional connectivity (FC) studies have demonstrated the overarching value of studying the brain and its disorders through the undirected weighted graph of fMRI correlation matrix. Most of the work with the FC, however, depends on the way the connectivity is computed, and further depends on the manual post-hoc analysis of the FC matrices. In this work we propose a deep learning architecture BrainGNN that learns the connectivity structure as part of learning to classify subjects. It simultaneously applies a graphical neural network to this learned graph and learns to select a sparse subset of brain regions important to the prediction task. We demonstrate the model's state-of-the-art classification performance on a schizophrenia fMRI dataset and demonstrate how introspection leads to disorder relevant findings. The graphs learned by the model exhibit strong class discrimination and the sparse subset of relevant regions are consistent with the schizophrenia literature.
* Accepted at Algorithms 2021, 14(3), 75
Multi network InfoMax: A pre-training method involving graph convolutional networks
Nov 01, 2021


Abstract:Discovering distinct features and their relations from data can help us uncover valuable knowledge crucial for various tasks, e.g., classification. In neuroimaging, these features could help to understand, classify, and possibly prevent brain disorders. Model introspection of highly performant overparameterized deep learning (DL) models could help find these features and relations. However, to achieve high-performance level DL models require numerous labeled training samples ($n$) rarely available in many fields. This paper presents a pre-training method involving graph convolutional/neural networks (GCNs/GNNs), based on maximizing mutual information between two high-level embeddings of an input sample. Many of the recently proposed pre-training methods pre-train one of many possible networks of an architecture. Since almost every DL model is an ensemble of multiple networks, we take our high-level embeddings from two different networks of a model --a convolutional and a graph network--. The learned high-level graph latent representations help increase performance for downstream graph classification tasks and bypass the need for a high number of labeled data samples. We apply our method to a neuroimaging dataset for classifying subjects into healthy control (HC) and schizophrenia (SZ) groups. Our experiments show that the pre-trained model significantly outperforms the non-pre-trained model and requires $50\%$ less data for similar performance.
Brain dynamics via Cumulative Auto-Regressive Self-Attention
Nov 01, 2021


Abstract:Multivariate dynamical processes can often be intuitively described by a weighted connectivity graph between components representing each individual time-series. Even a simple representation of this graph as a Pearson correlation matrix may be informative and predictive as demonstrated in the brain imaging literature. However, there is a consensus expectation that powerful graph neural networks (GNNs) should perform better in similar settings. In this work, we present a model that is considerably shallow than deep GNNs, yet outperforms them in predictive accuracy in a brain imaging application. Our model learns the autoregressive structure of individual time series and estimates directed connectivity graphs between the learned representations via a self-attention mechanism in an end-to-end fashion. The supervised training of the model as a classifier between patients and controls results in a model that generates directed connectivity graphs and highlights the components of the time-series that are predictive for each subject. We demonstrate our results on a functional neuroimaging dataset classifying schizophrenia patients and controls.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge