Usman Mahmood
Self-Supervised Mental Disorder Classifiers via Time Reversal
Nov 30, 2022Abstract:Data scarcity is a notable problem, especially in the medical domain, due to patient data laws. Therefore, efficient Pre-Training techniques could help in combating this problem. In this paper, we demonstrate that a model trained on the time direction of functional neuro-imaging data could help in any downstream task, for example, classifying diseases from healthy controls in fMRI data. We train a Deep Neural Network on Independent components derived from fMRI data using the Independent component analysis (ICA) technique. It learns time direction in the ICA-based data. This pre-trained model is further trained to classify brain disorders in different datasets. Through various experiments, we have shown that learning time direction helps a model learn some causal relation in fMRI data that helps in faster convergence, and consequently, the model generalizes well in downstream classification tasks even with fewer data records.
Fusion Subspace Clustering for Incomplete Data
May 22, 2022


Abstract:This paper introduces {\em fusion subspace clustering}, a novel method to learn low-dimensional structures that approximate large scale yet highly incomplete data. The main idea is to assign each datum to a subspace of its own, and minimize the distance between the subspaces of all data, so that subspaces of the same cluster get {\em fused} together. Our method allows low, high, and even full-rank data; it directly accounts for noise, and its sample complexity approaches the information-theoretic limit. In addition, our approach provides a natural model selection {\em clusterpath}, and a direct completion method. We give convergence guarantees, analyze computational complexity, and show through extensive experiments on real and synthetic data that our approach performs comparably to the state-of-the-art with complete data, and dramatically better if data is missing.
Deep Dynamic Effective Connectivity Estimation from Multivariate Time Series
Feb 16, 2022



Abstract:Recently, methods that represent data as a graph, such as graph neural networks (GNNs) have been successfully used to learn data representations and structures to solve classification and link prediction problems. The applications of such methods are vast and diverse, but most of the current work relies on the assumption of a static graph. This assumption does not hold for many highly dynamic systems, where the underlying connectivity structure is non-stationary and is mostly unobserved. Using a static model in these situations may result in sub-optimal performance. In contrast, modeling changes in graph structure with time can provide information about the system whose applications go beyond classification. Most work of this type does not learn effective connectivity and focuses on cross-correlation between nodes to generate undirected graphs. An undirected graph is unable to capture direction of an interaction which is vital in many fields, including neuroscience. To bridge this gap, we developed dynamic effective connectivity estimation via neural network training (DECENNT), a novel model to learn an interpretable directed and dynamic graph induced by the downstream classification/prediction task. DECENNT outperforms state-of-the-art (SOTA) methods on five different tasks and infers interpretable task-specific dynamic graphs. The dynamic graphs inferred from functional neuroimaging data align well with the existing literature and provide additional information. Additionally, the temporal attention module of DECENNT identifies time-intervals crucial for predictive downstream task from multivariate time series data.
A deep learning model for data-driven discovery of functional connectivity
Dec 07, 2021



Abstract:Functional connectivity (FC) studies have demonstrated the overarching value of studying the brain and its disorders through the undirected weighted graph of fMRI correlation matrix. Most of the work with the FC, however, depends on the way the connectivity is computed, and further depends on the manual post-hoc analysis of the FC matrices. In this work we propose a deep learning architecture BrainGNN that learns the connectivity structure as part of learning to classify subjects. It simultaneously applies a graphical neural network to this learned graph and learns to select a sparse subset of brain regions important to the prediction task. We demonstrate the model's state-of-the-art classification performance on a schizophrenia fMRI dataset and demonstrate how introspection leads to disorder relevant findings. The graphs learned by the model exhibit strong class discrimination and the sparse subset of relevant regions are consistent with the schizophrenia literature.
* Accepted at Algorithms 2021, 14(3), 75
Multi network InfoMax: A pre-training method involving graph convolutional networks
Nov 01, 2021


Abstract:Discovering distinct features and their relations from data can help us uncover valuable knowledge crucial for various tasks, e.g., classification. In neuroimaging, these features could help to understand, classify, and possibly prevent brain disorders. Model introspection of highly performant overparameterized deep learning (DL) models could help find these features and relations. However, to achieve high-performance level DL models require numerous labeled training samples ($n$) rarely available in many fields. This paper presents a pre-training method involving graph convolutional/neural networks (GCNs/GNNs), based on maximizing mutual information between two high-level embeddings of an input sample. Many of the recently proposed pre-training methods pre-train one of many possible networks of an architecture. Since almost every DL model is an ensemble of multiple networks, we take our high-level embeddings from two different networks of a model --a convolutional and a graph network--. The learned high-level graph latent representations help increase performance for downstream graph classification tasks and bypass the need for a high number of labeled data samples. We apply our method to a neuroimaging dataset for classifying subjects into healthy control (HC) and schizophrenia (SZ) groups. Our experiments show that the pre-trained model significantly outperforms the non-pre-trained model and requires $50\%$ less data for similar performance.
Brain dynamics via Cumulative Auto-Regressive Self-Attention
Nov 01, 2021


Abstract:Multivariate dynamical processes can often be intuitively described by a weighted connectivity graph between components representing each individual time-series. Even a simple representation of this graph as a Pearson correlation matrix may be informative and predictive as demonstrated in the brain imaging literature. However, there is a consensus expectation that powerful graph neural networks (GNNs) should perform better in similar settings. In this work, we present a model that is considerably shallow than deep GNNs, yet outperforms them in predictive accuracy in a brain imaging application. Our model learns the autoregressive structure of individual time series and estimates directed connectivity graphs between the learned representations via a self-attention mechanism in an end-to-end fashion. The supervised training of the model as a classifier between patients and controls results in a model that generates directed connectivity graphs and highlights the components of the time-series that are predictive for each subject. We demonstrate our results on a functional neuroimaging dataset classifying schizophrenia patients and controls.
Detecting Spurious Correlations with Sanity Tests for Artificial Intelligence Guided Radiology Systems
Mar 04, 2021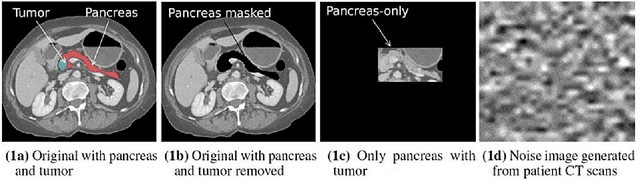
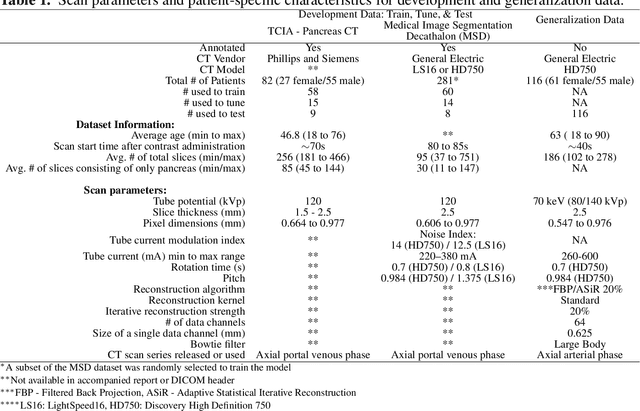
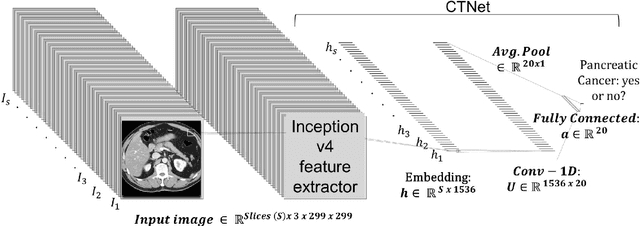
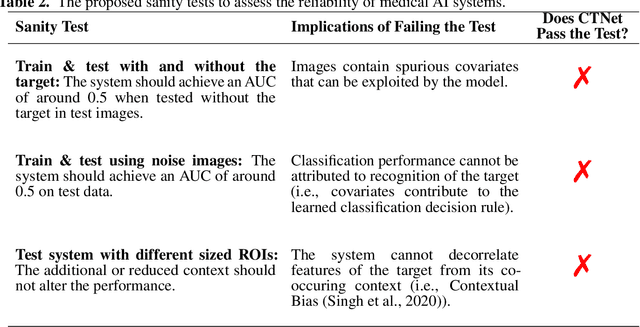
Abstract:Artificial intelligence (AI) has been successful at solving numerous problems in machine perception. In radiology, AI systems are rapidly evolving and show progress in guiding treatment decisions, diagnosing, localizing disease on medical images, and improving radiologists' efficiency. A critical component to deploying AI in radiology is to gain confidence in a developed system's efficacy and safety. The current gold standard approach is to conduct an analytical validation of performance on a generalization dataset from one or more institutions, followed by a clinical validation study of the system's efficacy during deployment. Clinical validation studies are time-consuming, and best practices dictate limited re-use of analytical validation data, so it is ideal to know ahead of time if a system is likely to fail analytical or clinical validation. In this paper, we describe a series of sanity tests to identify when a system performs well on development data for the wrong reasons. We illustrate the sanity tests' value by designing a deep learning system to classify pancreatic cancer seen in computed tomography scans.
Whole MILC: generalizing learned dynamics across tasks, datasets, and populations
Jul 29, 2020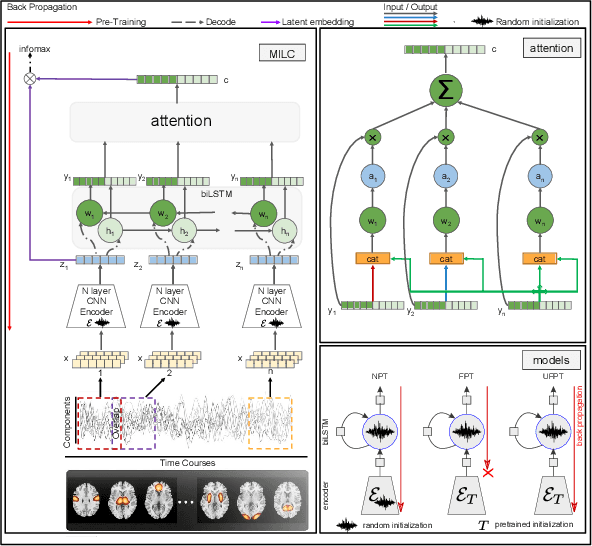
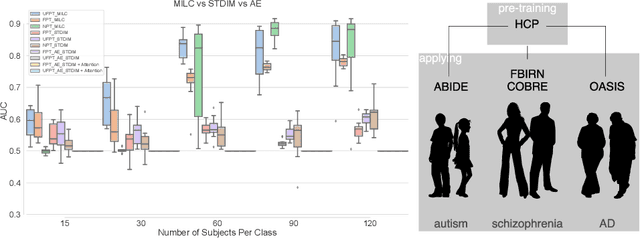
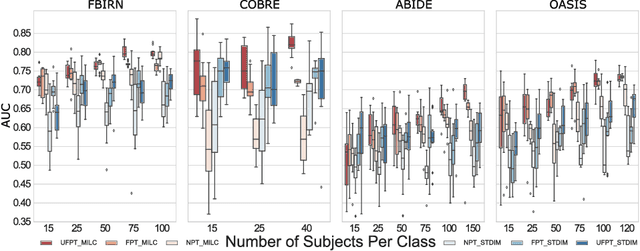
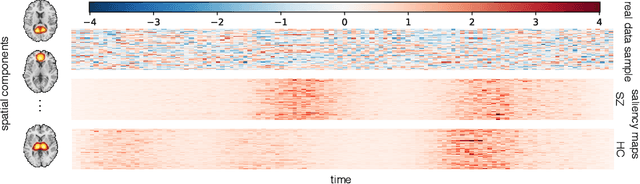
Abstract:Behavioral changes are the earliest signs of a mental disorder, but arguably, the dynamics of brain function gets affected even earlier. Subsequently, spatio-temporal structure of disorder-specific dynamics is crucial for early diagnosis and understanding the disorder mechanism. A common way of learning discriminatory features relies on training a classifier and evaluating feature importance. Classical classifiers, based on handcrafted features are quite powerful, but suffer the curse of dimensionality when applied to large input dimensions of spatio-temporal data. Deep learning algorithms could handle the problem and a model introspection could highlight discriminatory spatio-temporal regions but need way more samples to train. In this paper we present a novel self supervised training schema which reinforces whole sequence mutual information local to context (whole MILC). We pre-train the whole MILC model on unlabeled and unrelated healthy control data. We test our model on three different disorders (i) Schizophrenia (ii) Autism and (iii) Alzheimers and four different studies. Our algorithm outperforms existing self-supervised pre-training methods and provides competitive classification results to classical machine learning algorithms. Importantly, whole MILC enables attribution of subject diagnosis to specific spatio-temporal regions in the fMRI signal.
Transfer Learning of fMRI Dynamics
Nov 16, 2019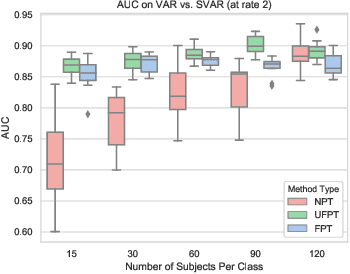
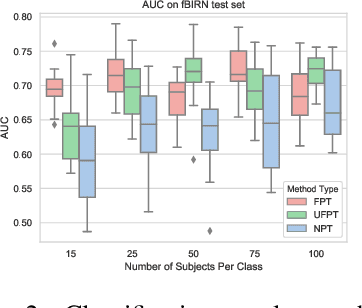
Abstract:As a mental disorder progresses, it may affect brain structure, but brain function expressed in brain dynamics is affected much earlier. Capturing the moment when brain dynamics express the disorder is crucial for early diagnosis. The traditional approach to this problem via training classifiers either proceeds from handcrafted features or requires large datasets to combat the $m>>n$ problem when a high dimensional fMRI volume only has a single label that carries learning signal. Large datasets may not be available for a study of each disorder, or rare disorder types or sub-populations may not warrant for them. In this paper, we demonstrate a self-supervised pre-training method that enables us to pre-train directly on fMRI dynamics of healthy control subjects and transfer the learning to much smaller datasets of schizophrenia. Not only we enable classification of disorder directly based on fMRI dynamics in small data but also significantly speed up the learning when possible. This is encouraging evidence of informative transfer learning across datasets and diagnostic categories.
Fusion Subspace Clustering: Full and Incomplete Data
Aug 02, 2018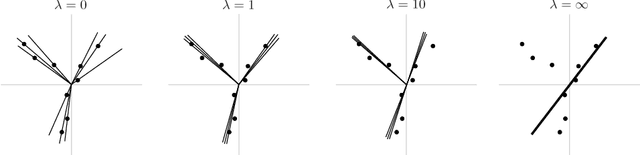
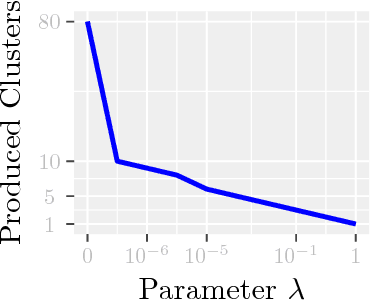
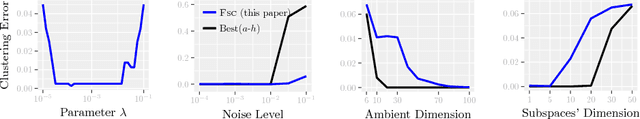
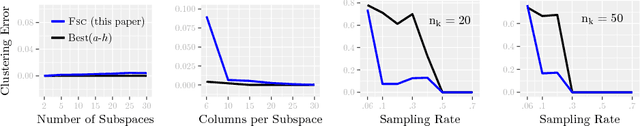
Abstract:Modern inference and learning often hinge on identifying low-dimensional structures that approximate large scale data. Subspace clustering achieves this through a union of linear subspaces. However, in contemporary applications data is increasingly often incomplete, rendering standard (full-data) methods inapplicable. On the other hand, existing incomplete-data methods present major drawbacks, like lifting an already high-dimensional problem, or requiring a super polynomial number of samples. Motivated by this, we introduce a new subspace clustering algorithm inspired by fusion penalties. The main idea is to permanently assign each datum to a subspace of its own, and minimize the distance between the subspaces of all data, so that subspaces of the same cluster get fused together. Our approach is entirely new to both, full and missing data, and unlike other methods, it directly allows noise, it requires no liftings, it allows low, high, and even full-rank data, it approaches optimal (information-theoretic) sampling rates, and it does not rely on other methods such as low-rank matrix completion to handle missing data. Furthermore, our extensive experiments on both real and synthetic data show that our approach performs comparably to the state-of-the-art with complete data, and dramatically better if data is missing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge