Sergey M. Plis
Tri-Institutional Center for Translational Research in Neuroimaging and Data Science, The Mind Research Network, Albuquerque, NM, USA
Guidelines For The Choice Of The Baseline in XAI Attribution Methods
Mar 25, 2025Abstract:Given the broad adoption of artificial intelligence, it is essential to provide evidence that AI models are reliable, trustable, and fair. To this end, the emerging field of eXplainable AI develops techniques to probe such requirements, counterbalancing the hype pushing the pervasiveness of this technology. Among the many facets of this issue, this paper focuses on baseline attribution methods, aiming at deriving a feature attribution map at the network input relying on a "neutral" stimulus usually called "baseline". The choice of the baseline is crucial as it determines the explanation of the network behavior. In this framework, this paper has the twofold goal of shedding light on the implications of the choice of the baseline and providing a simple yet effective method for identifying the best baseline for the task. To achieve this, we propose a decision boundary sampling method, since the baseline, by definition, lies on the decision boundary, which naturally becomes the search domain. Experiments are performed on synthetic examples and validated relying on state-of-the-art methods. Despite being limited to the experimental scope, this contribution is relevant as it offers clear guidelines and a simple proxy for baseline selection, reducing ambiguity and enhancing deep models' reliability and trust.
Spectral Introspection Identifies Group Training Dynamics in Deep Neural Networks for Neuroimaging
Jun 17, 2024
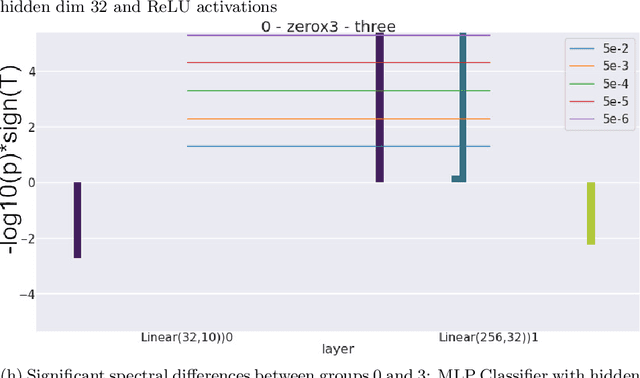
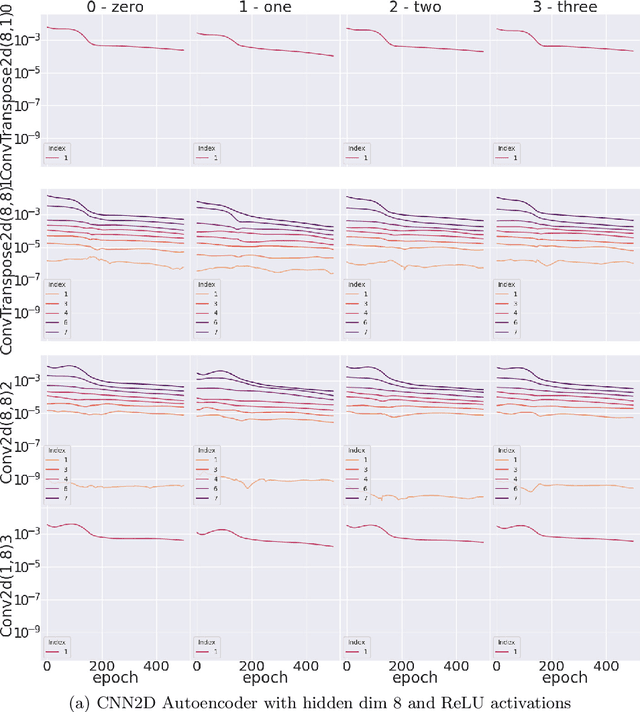
Abstract:Neural networks, whice have had a profound effect on how researchers study complex phenomena, do so through a complex, nonlinear mathematical structure which can be difficult for human researchers to interpret. This obstacle can be especially salient when researchers want to better understand the emergence of particular model behaviors such as bias, overfitting, overparametrization, and more. In Neuroimaging, the understanding of how such phenomena emerge is fundamental to preventing and informing users of the potential risks involved in practice. In this work, we present a novel introspection framework for Deep Learning on Neuroimaging data, which exploits the natural structure of gradient computations via the singular value decomposition of gradient components during reverse-mode auto-differentiation. Unlike post-hoc introspection techniques, which require fully-trained models for evaluation, our method allows for the study of training dynamics on the fly, and even more interestingly, allow for the decomposition of gradients based on which samples belong to particular groups of interest. We demonstrate how the gradient spectra for several common deep learning models differ between schizophrenia and control participants from the COBRE study, and illustrate how these trajectories may reveal specific training dynamics helpful for further analysis.
Low-Rank Learning by Design: the Role of Network Architecture and Activation Linearity in Gradient Rank Collapse
Feb 09, 2024
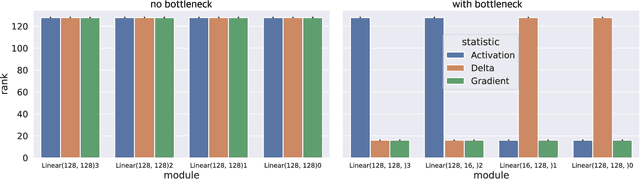
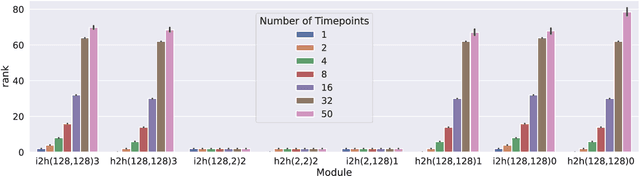
Abstract:Our understanding of learning dynamics of deep neural networks (DNNs) remains incomplete. Recent research has begun to uncover the mathematical principles underlying these networks, including the phenomenon of "Neural Collapse", where linear classifiers within DNNs converge to specific geometrical structures during late-stage training. However, the role of geometric constraints in learning extends beyond this terminal phase. For instance, gradients in fully-connected layers naturally develop a low-rank structure due to the accumulation of rank-one outer products over a training batch. Despite the attention given to methods that exploit this structure for memory saving or regularization, the emergence of low-rank learning as an inherent aspect of certain DNN architectures has been under-explored. In this paper, we conduct a comprehensive study of gradient rank in DNNs, examining how architectural choices and structure of the data effect gradient rank bounds. Our theoretical analysis provides these bounds for training fully-connected, recurrent, and convolutional neural networks. We also demonstrate, both theoretically and empirically, how design choices like activation function linearity, bottleneck layer introduction, convolutional stride, and sequence truncation influence these bounds. Our findings not only contribute to the understanding of learning dynamics in DNNs, but also provide practical guidance for deep learning engineers to make informed design decisions.
Looking deeper into interpretable deep learning in neuroimaging: a comprehensive survey
Jul 14, 2023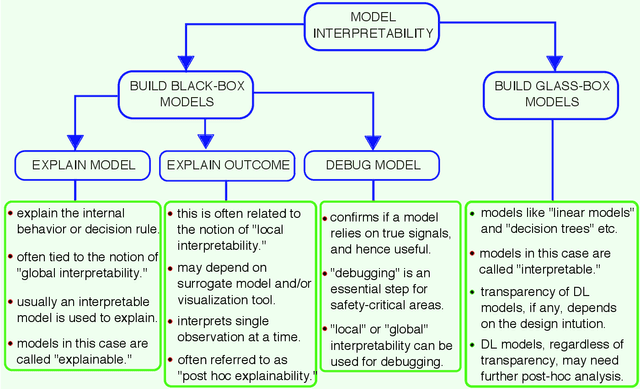
Abstract:Deep learning (DL) models have been popular due to their ability to learn directly from the raw data in an end-to-end paradigm, alleviating the concern of a separate error-prone feature extraction phase. Recent DL-based neuroimaging studies have also witnessed a noticeable performance advancement over traditional machine learning algorithms. But the challenges of deep learning models still exist because of the lack of transparency in these models for their successful deployment in real-world applications. In recent years, Explainable AI (XAI) has undergone a surge of developments mainly to get intuitions of how the models reached the decisions, which is essential for safety-critical domains such as healthcare, finance, and law enforcement agencies. While the interpretability domain is advancing noticeably, researchers are still unclear about what aspect of model learning a post hoc method reveals and how to validate its reliability. This paper comprehensively reviews interpretable deep learning models in the neuroimaging domain. Firstly, we summarize the current status of interpretability resources in general, focusing on the progression of methods, associated challenges, and opinions. Secondly, we discuss how multiple recent neuroimaging studies leveraged model interpretability to capture anatomical and functional brain alterations most relevant to model predictions. Finally, we discuss the limitations of the current practices and offer some valuable insights and guidance on how we can steer our future research directions to make deep learning models substantially interpretable and thus advance scientific understanding of brain disorders.
Self-supervised multimodal neuroimaging yields predictive representations for a spectrum of Alzheimer's phenotypes
Sep 07, 2022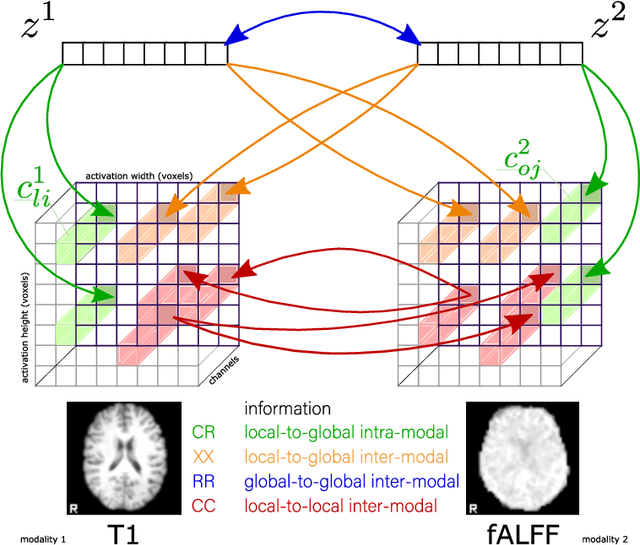
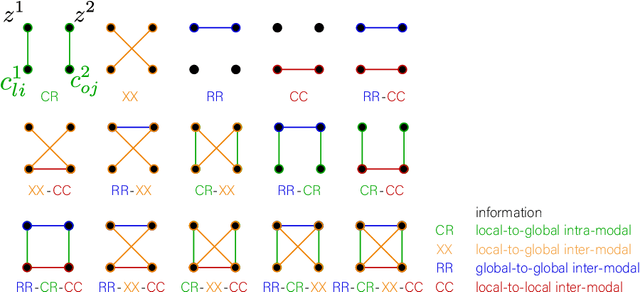
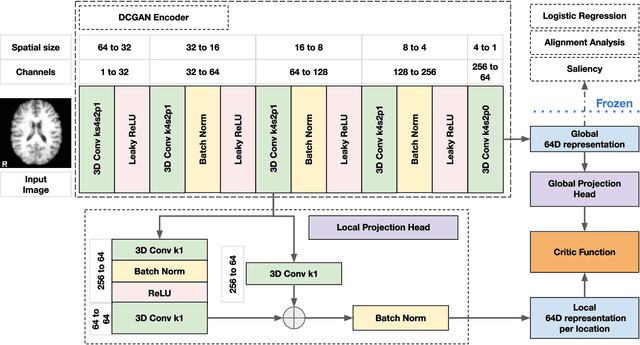
Abstract:Recent neuroimaging studies that focus on predicting brain disorders via modern machine learning approaches commonly include a single modality and rely on supervised over-parameterized models.However, a single modality provides only a limited view of the highly complex brain. Critically, supervised models in clinical settings lack accurate diagnostic labels for training. Coarse labels do not capture the long-tailed spectrum of brain disorder phenotypes, which leads to a loss of generalizability of the model that makes them less useful in diagnostic settings. This work presents a novel multi-scale coordinated framework for learning multiple representations from multimodal neuroimaging data. We propose a general taxonomy of informative inductive biases to capture unique and joint information in multimodal self-supervised fusion. The taxonomy forms a family of decoder-free models with reduced computational complexity and a propensity to capture multi-scale relationships between local and global representations of the multimodal inputs. We conduct a comprehensive evaluation of the taxonomy using functional and structural magnetic resonance imaging (MRI) data across a spectrum of Alzheimer's disease phenotypes and show that self-supervised models reveal disorder-relevant brain regions and multimodal links without access to the labels during pre-training. The proposed multimodal self-supervised learning yields representations with improved classification performance for both modalities. The concomitant rich and flexible unsupervised deep learning framework captures complex multimodal relationships and provides predictive performance that meets or exceeds that of a more narrow supervised classification analysis. We present elaborate quantitative evidence of how this framework can significantly advance our search for missing links in complex brain disorders.
Algorithm-Agnostic Explainability for Unsupervised Clustering
May 17, 2021
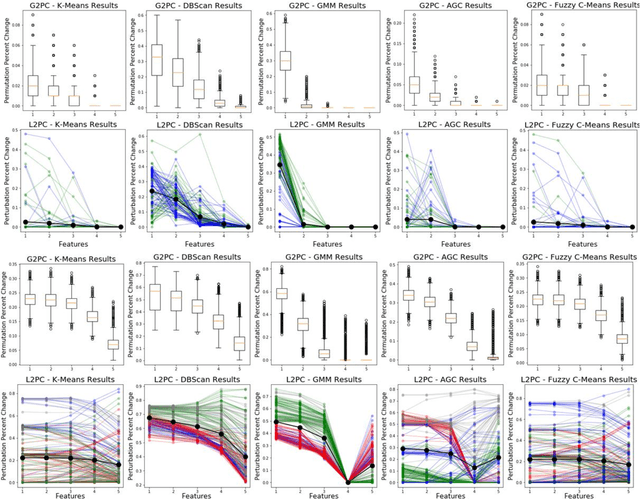
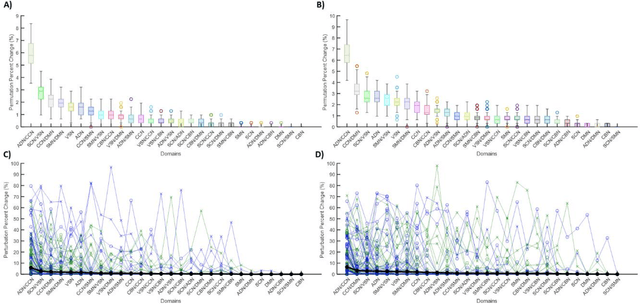
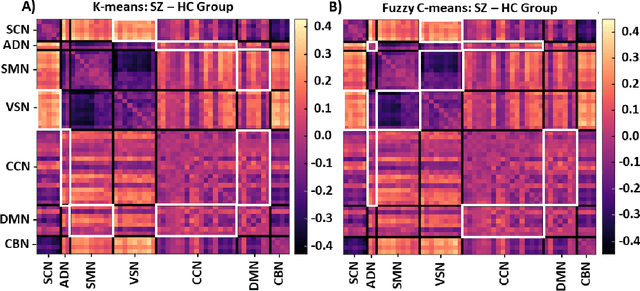
Abstract:Supervised machine learning explainability has greatly expanded in recent years. However, the field of unsupervised clustering explainability has lagged behind. Here, we, to the best of our knowledge, demonstrate for the first time how model-agnostic methods for supervised machine learning explainability can be adapted to provide algorithm-agnostic unsupervised clustering explainability. We present two novel algorithm-agnostic explainability methods, global permutation percent change (G2PC) feature importance and local perturbation percent change (L2PC) feature importance, that can provide insight into many clustering methods on a global level by identifying the relative importance of features to a clustering algorithm and on a local level by identifying the relative importance of features to the clustering of individual samples. We demonstrate the utility of the methods for explaining five popular clustering algorithms on low-dimensional, ground-truth synthetic datasets and on high-dimensional functional network connectivity (FNC) data extracted from a resting state functional magnetic resonance imaging (rs-fMRI) dataset of 151 subjects with schizophrenia (SZ) and 160 healthy controls (HC). Our proposed explainability methods robustly identify the relative importance of features across multiple clustering methods and could facilitate new insights into many applications. We hope that this study will greatly accelerate the development of the field of clustering explainability.
Tasting the cake: evaluating self-supervised generalization on out-of-distribution multimodal MRI data
Apr 20, 2021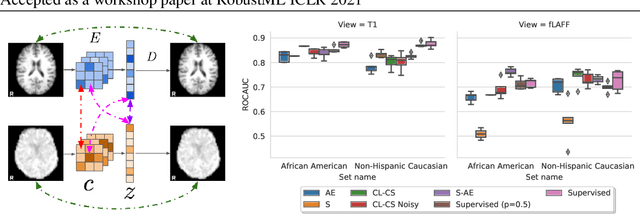
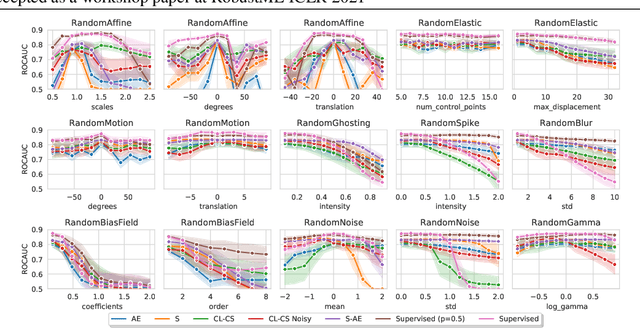
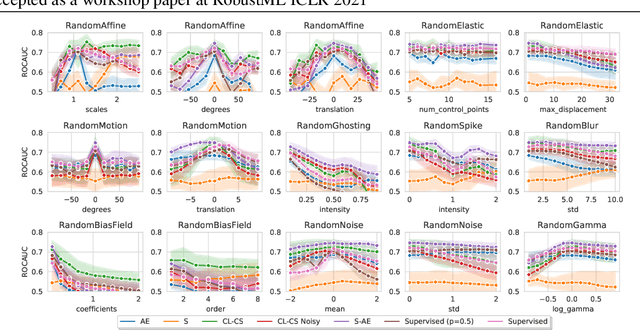

Abstract:Self-supervised learning has enabled significant improvements on natural image benchmarks. However, there is less work in the medical imaging domain in this area. The optimal models have not yet been determined among the various options. Moreover, little work has evaluated the current applicability limits of novel self-supervised methods. In this paper, we evaluate a range of current contrastive self-supervised methods on out-of-distribution generalization in order to evaluate their applicability to medical imaging. We show that self-supervised models are not as robust as expected based on their results in natural imaging benchmarks and can be outperformed by supervised learning with dropout. We also show that this behavior can be countered with extensive augmentation. Our results highlight the need for out-of-distribution generalization standards and benchmarks to adopt the self-supervised methods in the medical imaging community.
Efficient Distributed Auto-Differentiation
Feb 22, 2021
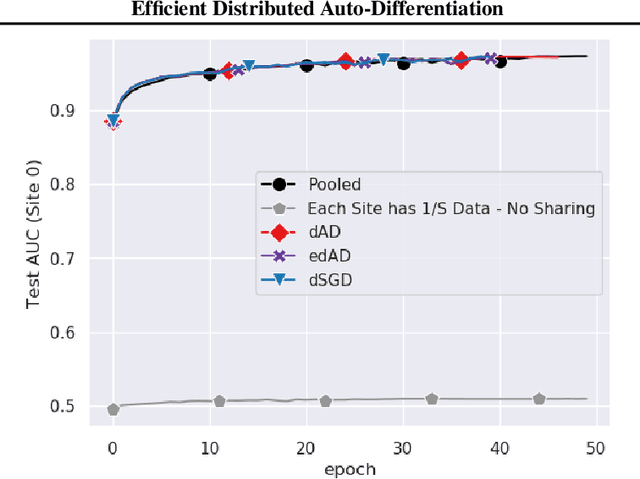
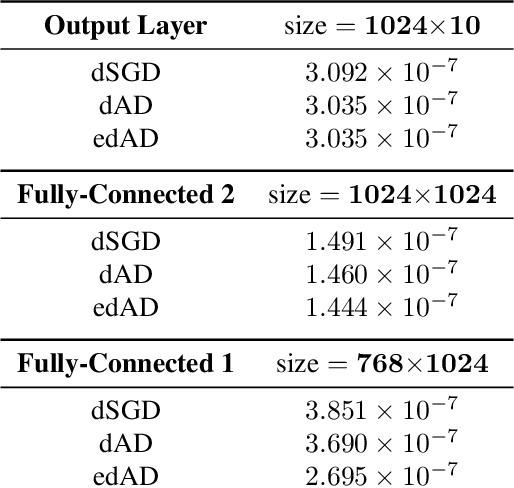

Abstract:Although distributed machine learning has opened up numerous frontiers of research, the separation of large models across different devices, nodes, and sites can invite significant communication overhead, making reliable training difficult. The focus on gradients as the primary shared statistic during training has led to a number of intuitive algorithms for distributed deep learning; however, gradient-based algorithms for training large deep neural networks (DNNs) are communication-heavy, often requiring additional modifications via sparsity constraints, compression, quantization, and other similar approaches, to lower bandwidth. We introduce a surprisingly simple statistic for training distributed DNNs that is more communication-friendly than the gradient. The error backpropagation process can be modified to share these smaller intermediate values instead of the gradient, reducing communication overhead with no impact on accuracy. The process provides the flexibility of averaging gradients during backpropagation, enabling novel flexible training schemas while leaving room for further bandwidth reduction via existing gradient compression methods. Finally, consideration of the matrices used to compute the gradient inspires a new approach to compression via structured power iterations, which can not only reduce bandwidth but also enable introspection into distributed training dynamics, without significant performance loss.
Taxonomy of multimodal self-supervised representation learning
Dec 29, 2020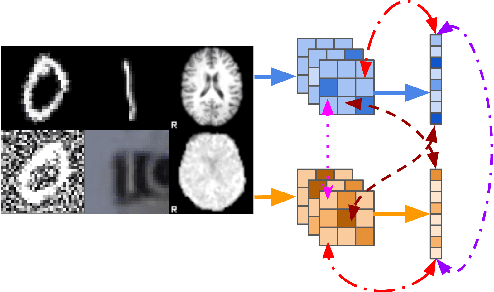
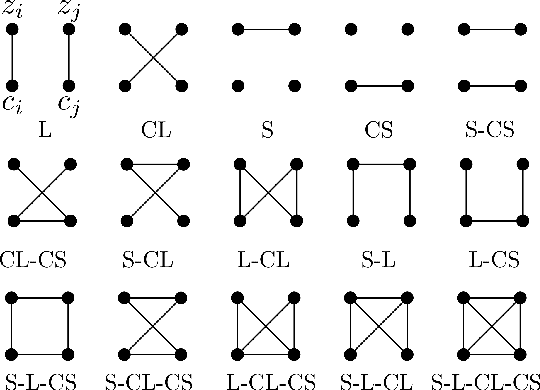
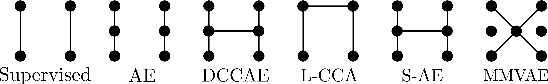
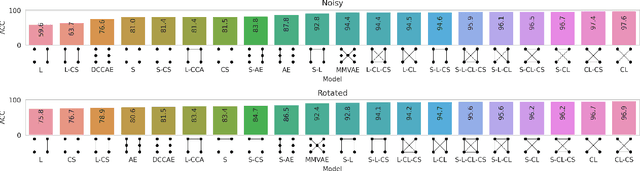
Abstract:Sensory input from multiple sources is crucial for robust and coherent human perception. Different sources contribute complementary explanatory factors and get combined based on factors they share. This system motivated the design of powerful unsupervised representation-learning algorithms. In this paper, we unify recent work on multimodal self-supervised learning under a single framework. Observing that most self-supervised methods optimize similarity metrics between a set of model components, we propose a taxonomy of all reasonable ways to organize this process. We empirically show on two versions of multimodal MNIST and a multimodal brain imaging dataset that (1) multimodal contrastive learning has significant benefits over its unimodal counterpart, (2) the specific composition of multiple contrastive objectives is critical to performance on a downstream task, (3) maximization of the similarity between representations has a regularizing effect on a neural network, which sometimes can lead to reduced downstream performance but still can reveal multimodal relations. Consequently, we outperform previous unsupervised encoder-decoder methods based on CCA or variational mixtures MMVAE on various datasets on linear evaluation protocol.
On self-supervised multi-modal representation learning: An application to Alzheimer's disease
Dec 25, 2020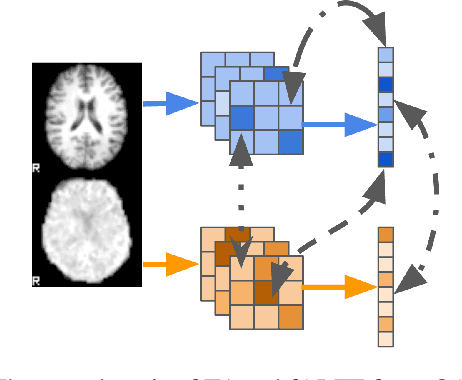
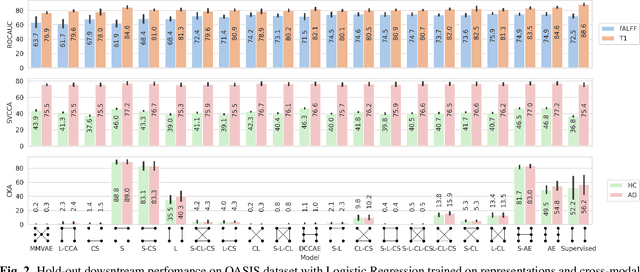
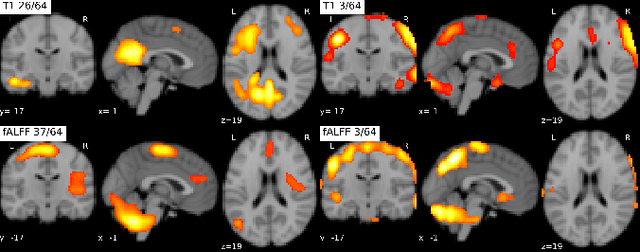
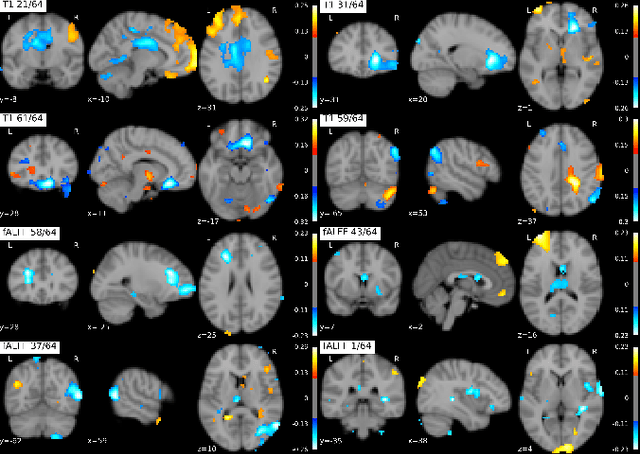
Abstract:Introspection of deep supervised predictive models trained on functional and structural brain imaging may uncover novel markers of Alzheimer's disease (AD). However, supervised training is prone to learning from spurious features (shortcut learning) impairing its value in the discovery process. Deep unsupervised and, recently, contrastive self-supervised approaches, not biased to classification, are better candidates for the task. Their multimodal options specifically offer additional regularization via modality interactions. In this paper, we introduce a way to exhaustively consider multimodal architectures for contrastive self-supervised fusion of fMRI and MRI of AD patients and controls. We show that this multimodal fusion results in representations that improve the results of the downstream classification for both modalities. We investigate the fused self-supervised features projected into the brain space and introduce a numerically stable way to do so.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge