On self-supervised multi-modal representation learning: An application to Alzheimer's disease
Paper and Code
Dec 25, 2020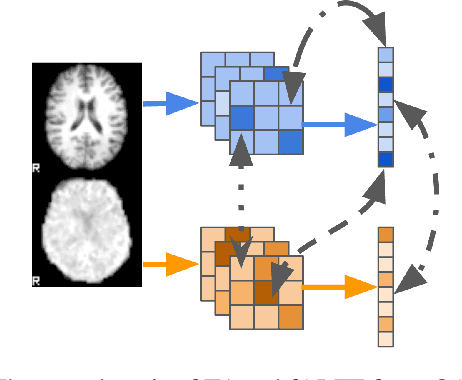
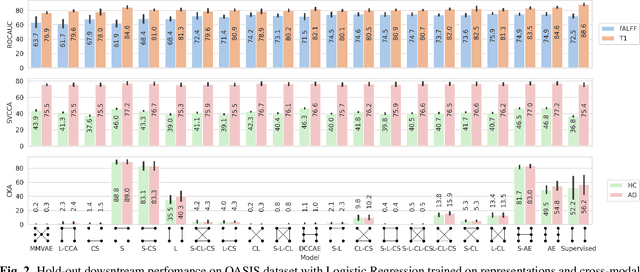
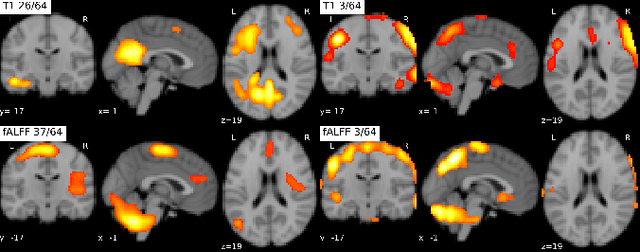
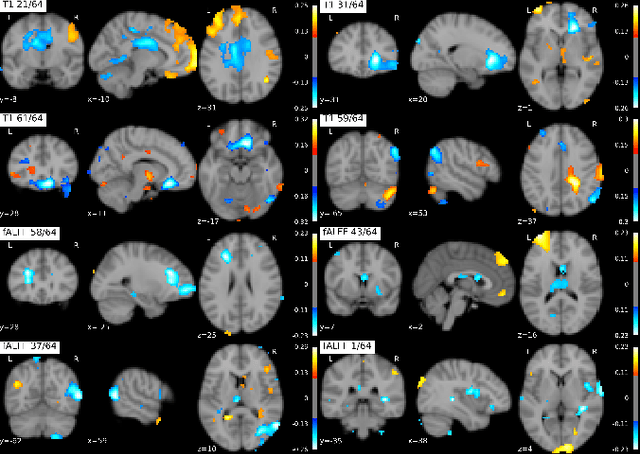
Introspection of deep supervised predictive models trained on functional and structural brain imaging may uncover novel markers of Alzheimer's disease (AD). However, supervised training is prone to learning from spurious features (shortcut learning) impairing its value in the discovery process. Deep unsupervised and, recently, contrastive self-supervised approaches, not biased to classification, are better candidates for the task. Their multimodal options specifically offer additional regularization via modality interactions. In this paper, we introduce a way to exhaustively consider multimodal architectures for contrastive self-supervised fusion of fMRI and MRI of AD patients and controls. We show that this multimodal fusion results in representations that improve the results of the downstream classification for both modalities. We investigate the fused self-supervised features projected into the brain space and introduce a numerically stable way to do so.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge