Noah Lewis
I/O in Machine Learning Applications on HPC Systems: A 360-degree Survey
Apr 16, 2024



Abstract:High-Performance Computing (HPC) systems excel in managing distributed workloads, and the growing interest in Artificial Intelligence (AI) has resulted in a surge in demand for faster methods of Machine Learning (ML) model training and inference. In the past, research on HPC I/O focused on optimizing the underlying storage system for modeling and simulation applications and checkpointing the results, causing writes to be the dominant I/O operation. These applications typically access large portions of the data written by simulations or experiments. ML workloads, in contrast, perform small I/O reads spread across a large number of random files. This shift of I/O access patterns poses several challenges to HPC storage systems. In this paper, we survey I/O in ML applications on HPC systems, and target literature within a 6-year time window from 2019 to 2024. We provide an overview of the common phases of ML, review available profilers and benchmarks, examine the I/O patterns encountered during ML training, explore I/O optimizations utilized in modern ML frameworks and proposed in recent literature, and lastly, present gaps requiring further R&D. We seek to summarize the common practices used in accessing data by ML applications and expose research gaps that could spawn further R&D.
CommsVAE: Learning the brain's macroscale communication dynamics using coupled sequential VAEs
Oct 07, 2022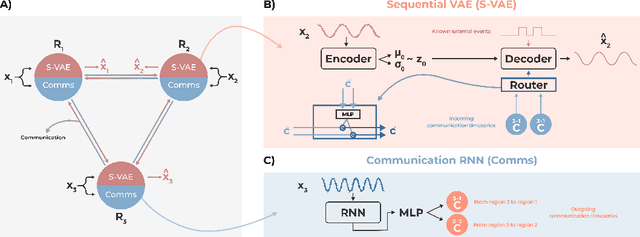
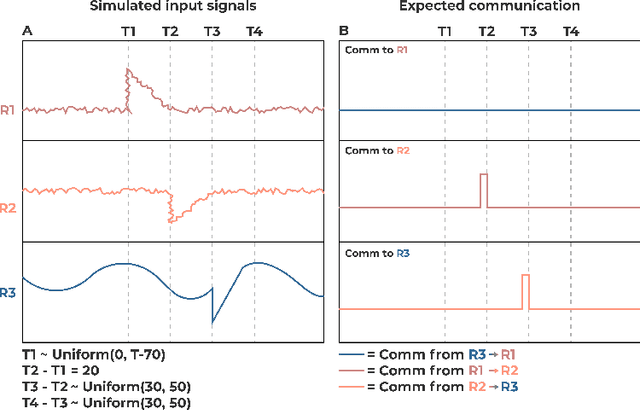
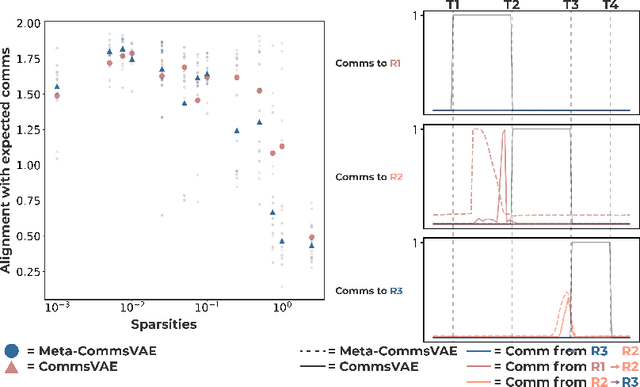
Abstract:Communication within or between complex systems is commonplace in the natural sciences and fields such as graph neural networks. The brain is a perfect example of such a complex system, where communication between brain regions is constantly being orchestrated. To analyze communication, the brain is often split up into anatomical regions that each perform certain computations. These regions must interact and communicate with each other to perform tasks and support higher-level cognition. On a macroscale, these regions communicate through signal propagation along the cortex and along white matter tracts over longer distances. When and what types of signals are communicated over time is an unsolved problem and is often studied using either functional or structural data. In this paper, we propose a non-linear generative approach to communication from functional data. We address three issues with common connectivity approaches by explicitly modeling the directionality of communication, finding communication at each timestep, and encouraging sparsity. To evaluate our model, we simulate temporal data that has sparse communication between nodes embedded in it and show that our model can uncover the expected communication dynamics. Subsequently, we apply our model to temporal neural data from multiple tasks and show that our approach models communication that is more specific to each task. The specificity of our method means it can have an impact on the understanding of psychiatric disorders, which are believed to be related to highly specific communication between brain regions compared to controls. In sum, we propose a general model for dynamic communication learning on graphs, and show its applicability to a subfield of the natural sciences, with potential widespread scientific impact.
Geometrically Guided Integrated Gradients
Jun 16, 2022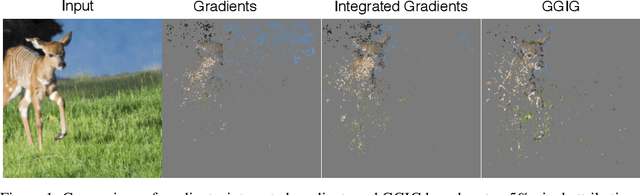
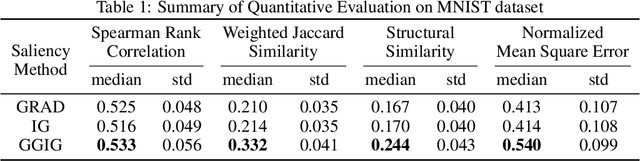
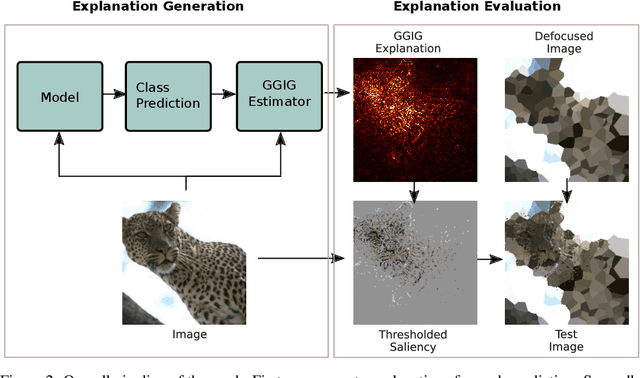
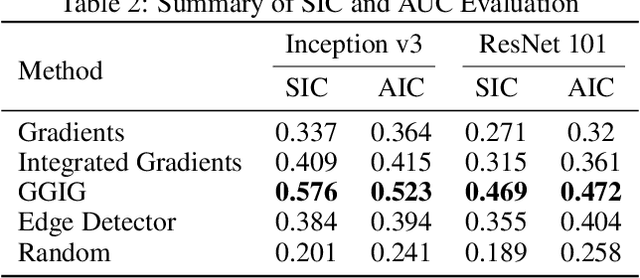
Abstract:Interpretability methods for deep neural networks mainly focus on the sensitivity of the class score with respect to the original or perturbed input, usually measured using actual or modified gradients. Some methods also use a model-agnostic approach to understanding the rationale behind every prediction. In this paper, we argue and demonstrate that local geometry of the model parameter space relative to the input can also be beneficial for improved post-hoc explanations. To achieve this goal, we introduce an interpretability method called "geometrically-guided integrated gradients" that builds on top of the gradient calculation along a linear path as traditionally used in integrated gradient methods. However, instead of integrating gradient information, our method explores the model's dynamic behavior from multiple scaled versions of the input and captures the best possible attribution for each input. We demonstrate through extensive experiments that the proposed approach outperforms vanilla and integrated gradients in subjective and quantitative assessment. We also propose a "model perturbation" sanity check to complement the traditionally used "model randomization" test.
Spatio-temporally separable non-linear latent factor learning: an application to somatomotor cortex fMRI data
May 26, 2022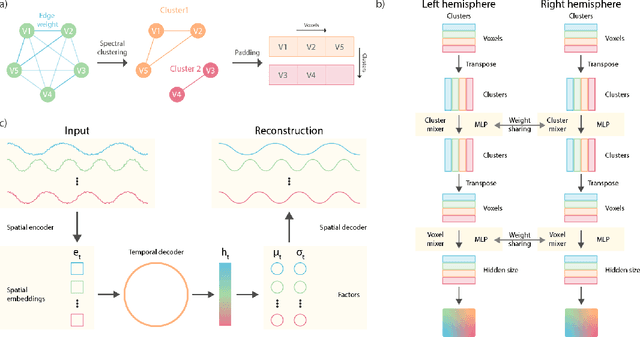
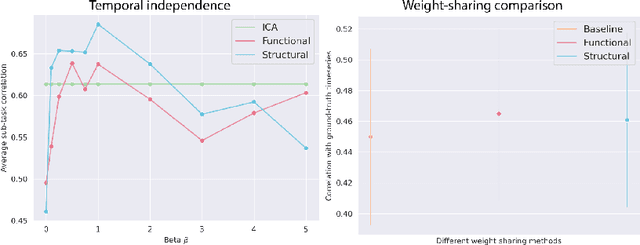
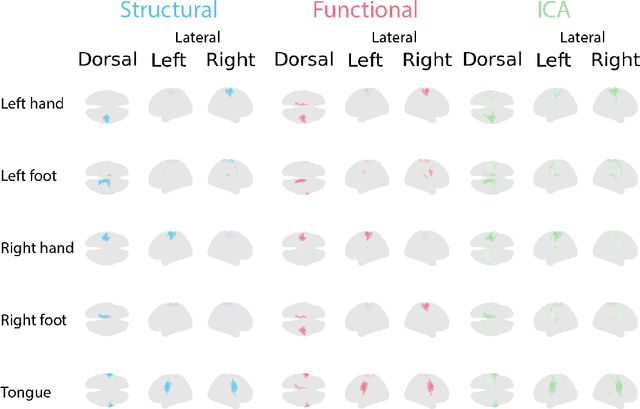
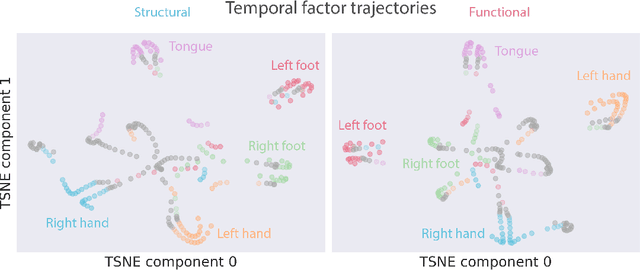
Abstract:Functional magnetic resonance imaging (fMRI) data contain complex spatiotemporal dynamics, thus researchers have developed approaches that reduce the dimensionality of the signal while extracting relevant and interpretable dynamics. Models of fMRI data that can perform whole-brain discovery of dynamical latent factors are understudied. The benefits of approaches such as linear independent component analysis models have been widely appreciated, however, nonlinear extensions of these models present challenges in terms of identification. Deep learning methods provide a way forward, but new methods for efficient spatial weight-sharing are critical to deal with the high dimensionality of the data and the presence of noise. Our approach generalizes weight sharing to non-Euclidean neuroimaging data by first performing spectral clustering based on the structural and functional similarity between voxels. The spectral clusters and their assignments can then be used as patches in an adapted multi-layer perceptron (MLP)-mixer model to share parameters among input points. To encourage temporally independent latent factors, we use an additional total correlation term in the loss. Our approach is evaluated on data with multiple motor sub-tasks to assess whether the model captures disentangled latent factors that correspond to each sub-task. Then, to assess the latent factors we find further, we compare the spatial location of each latent factor to the motor homunculus. Finally, we show that our approach captures task effects better than the current gold standard of source signal separation, independent component analysis (ICA).
Fusing multimodal neuroimaging data with a variational autoencoder
May 03, 2021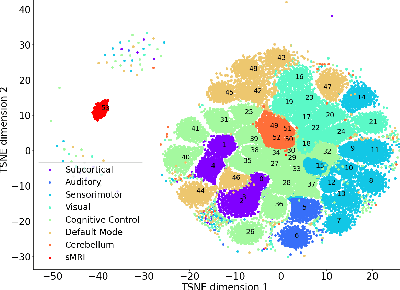
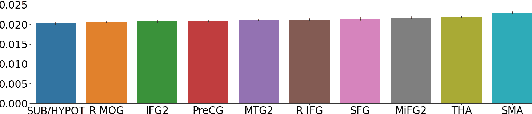
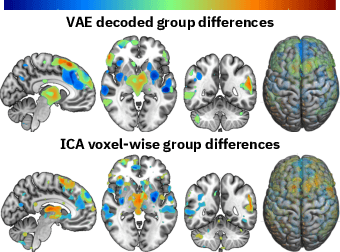
Abstract:Neuroimaging studies often involve the collection of multiple data modalities. These modalities contain both shared and mutually exclusive information about the brain. This work aims at finding a scalable and interpretable method to fuse the information of multiple neuroimaging modalities using a variational autoencoder (VAE). To provide an initial assessment, this work evaluates the representations that are learned using a schizophrenia classification task. A support vector machine trained on the representations achieves an area under the curve for the classifier's receiver operating characteristic (ROC-AUC) of 0.8610.
Whole MILC: generalizing learned dynamics across tasks, datasets, and populations
Jul 29, 2020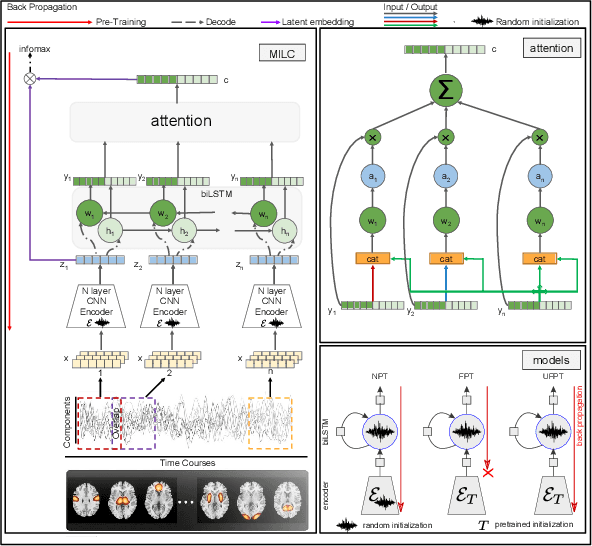
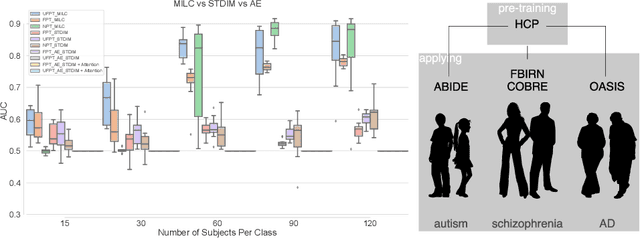
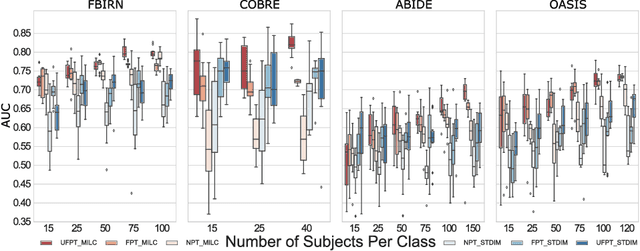
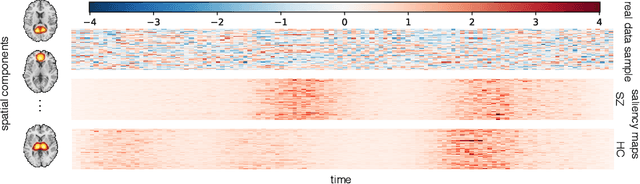
Abstract:Behavioral changes are the earliest signs of a mental disorder, but arguably, the dynamics of brain function gets affected even earlier. Subsequently, spatio-temporal structure of disorder-specific dynamics is crucial for early diagnosis and understanding the disorder mechanism. A common way of learning discriminatory features relies on training a classifier and evaluating feature importance. Classical classifiers, based on handcrafted features are quite powerful, but suffer the curse of dimensionality when applied to large input dimensions of spatio-temporal data. Deep learning algorithms could handle the problem and a model introspection could highlight discriminatory spatio-temporal regions but need way more samples to train. In this paper we present a novel self supervised training schema which reinforces whole sequence mutual information local to context (whole MILC). We pre-train the whole MILC model on unlabeled and unrelated healthy control data. We test our model on three different disorders (i) Schizophrenia (ii) Autism and (iii) Alzheimers and four different studies. Our algorithm outperforms existing self-supervised pre-training methods and provides competitive classification results to classical machine learning algorithms. Importantly, whole MILC enables attribution of subject diagnosis to specific spatio-temporal regions in the fMRI signal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge