Jing Sui
Cross-Modal Synthesis of Structural MRI and Functional Connectivity Networks via Conditional ViT-GANs
Sep 15, 2023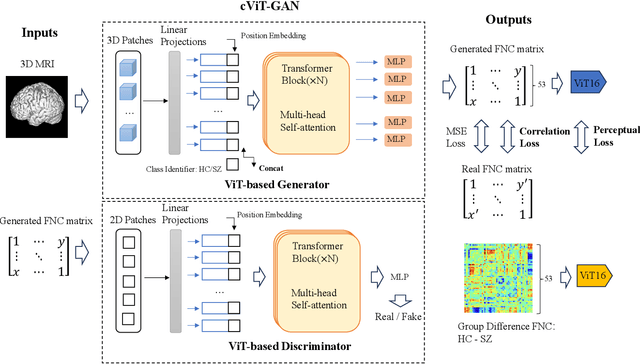
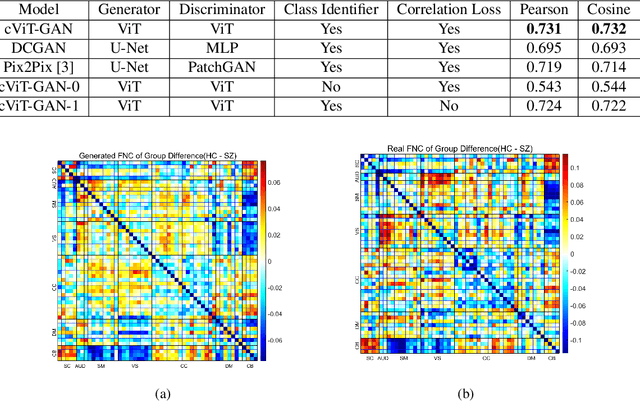
Abstract:The cross-modal synthesis between structural magnetic resonance imaging (sMRI) and functional network connectivity (FNC) is a relatively unexplored area in medical imaging, especially with respect to schizophrenia. This study employs conditional Vision Transformer Generative Adversarial Networks (cViT-GANs) to generate FNC data based on sMRI inputs. After training on a comprehensive dataset that included both individuals with schizophrenia and healthy control subjects, our cViT-GAN model effectively synthesized the FNC matrix for each subject, and then formed a group difference FNC matrix, obtaining a Pearson correlation of 0.73 with the actual FNC matrix. In addition, our FNC visualization results demonstrate significant correlations in particular subcortical brain regions, highlighting the model's capability of capturing detailed structural-functional associations. This performance distinguishes our model from conditional CNN-based GAN alternatives such as Pix2Pix. Our research is one of the first attempts to link sMRI and FNC synthesis, setting it apart from other cross-modal studies that concentrate on T1- and T2-weighted MR images or the fusion of MRI and CT scans.
Meta-modal Information Flow: A Method for Capturing Multimodal Modular Disconnectivity in Schizophrenia
Jan 06, 2020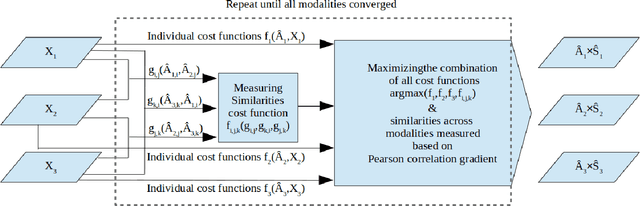
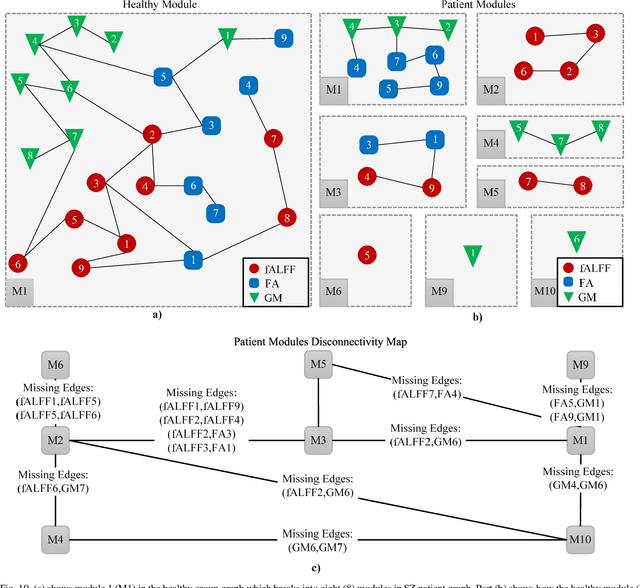
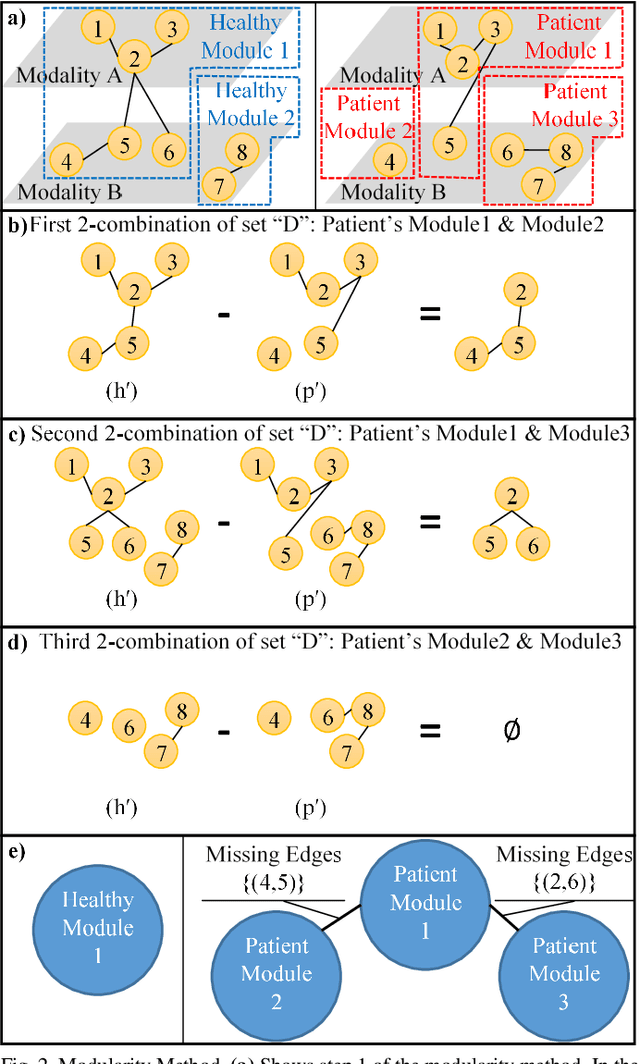
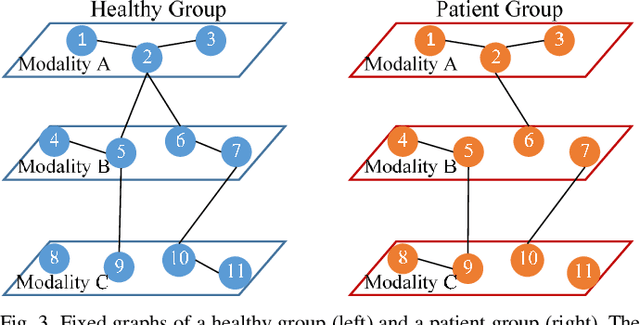
Abstract:Objective: Multimodal measurements of the same phenomena provide complementary information and highlight different perspectives, albeit each with their own limitations. A focus on a single modality may lead to incorrect inferences, which is especially important when a studied phenomenon is a disease. In this paper, we introduce a method that takes advantage of multimodal data in addressing the hypotheses of disconnectivity and dysfunction within schizophrenia (SZ). Methods: We start with estimating and visualizing links within and among extracted multimodal data features using a Gaussian graphical model (GGM). We then propose a modularity-based method that can be applied to the GGM to identify links that are associated with mental illness across a multimodal data set. Through simulation and real data, we show our approach reveals important information about disease-related network disruptions that are missed with a focus on a single modality. We use functional MRI (fMRI), diffusion MRI (dMRI), and structural MRI (sMRI) to compute the fractional amplitude of low frequency fluctuations (fALFF), fractional anisotropy (FA), and gray matter (GM) concentration maps. These three modalities are analyzed using our modularity method. Results: Our results show missing links that are only captured by the cross-modal information that may play an important role in disconnectivity between the components. Conclusion: We identified multimodal (fALFF, FA and GM) disconnectivity in the default mode network area in patients with SZ, which would not have been detectable in a single modality. Significance: The proposed approach provides an important new tool for capturing information that is distributed among multiple imaging modalities.
Deep Chronnectome Learning via Full Bidirectional Long Short-Term Memory Networks for MCI Diagnosis
Aug 30, 2018



Abstract:Brain functional connectivity (FC) extracted from resting-state fMRI (RS-fMRI) has become a popular approach for disease diagnosis, where discriminating subjects with mild cognitive impairment (MCI) from normal controls (NC) is still one of the most challenging problems. Dynamic functional connectivity (dFC), consisting of time-varying spatiotemporal dynamics, may characterize "chronnectome" diagnostic information for improving MCI classification. However, most of the current dFC studies are based on detecting discrete major brain status via spatial clustering, which ignores rich spatiotemporal dynamics contained in such chronnectome. We propose Deep Chronnectome Learning for exhaustively mining the comprehensive information, especially the hidden higher-level features, i.e., the dFC time series that may add critical diagnostic power for MCI classification. To this end, we devise a new Fully-connected Bidirectional Long Short-Term Memory Network (Full-BiLSTM) to effectively learn the periodic brain status changes using both past and future information for each brief time segment and then fuse them to form the final output. We have applied our method to a rigorously built large-scale multi-site database (i.e., with 164 data from NCs and 330 from MCIs, which can be further augmented by 25 folds). Our method outperforms other state-of-the-art approaches with an accuracy of 73.6% under solid cross-validations. We also made extensive comparisons among multiple variants of LSTM models. The results suggest high feasibility of our method with promising value also for other brain disorder diagnoses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge