Dong Hye Ye
KOCOBrain: Kuramoto-Guided Graph Network for Uncovering Structure-Function Coupling in Adolescent Prenatal Drug Exposure
Jan 16, 2026Abstract:Exposure to psychoactive substances during pregnancy, such as cannabis, can disrupt neurodevelopment and alter large-scale brain networks, yet identifying their neural signatures remains challenging. We introduced KOCOBrain: KuramotO COupled Brain Graph Network; a unified graph neural network framework that integrates structural and functional connectomes via Kuramoto-based phase dynamics and cognition-aware attention. The Kuramoto layer models neural synchronization over anatomical connections, generating phase-informed embeddings that capture structure-function coupling, while cognitive scores modulate information routing in a subject-specific manner followed by a joint objective enhancing robustness under class imbalance scenario. Applied to the ABCD cohort, KOCOBrain improved prenatal drug exposure prediction over relevant baselines and revealed interpretable structure-function patterns that reflect disrupted brain network coordination associated with early exposure.
Self-learned representation-guided latent diffusion model for breast cancer classification in deep ultraviolet whole surface images
Jan 16, 2026Abstract:Breast-Conserving Surgery (BCS) requires precise intraoperative margin assessment to preserve healthy tissue. Deep Ultraviolet Fluorescence Scanning Microscopy (DUV-FSM) offers rapid, high-resolution surface imaging for this purpose; however, the scarcity of annotated DUV data hinders the training of robust deep learning models. To address this, we propose an Self-Supervised Learning (SSL)-guided Latent Diffusion Model (LDM) to generate high-quality synthetic training patches. By guiding the LDM with embeddings from a fine-tuned DINO teacher, we inject rich semantic details of cellular structures into the synthetic data. We combine real and synthetic patches to fine-tune a Vision Transformer (ViT), utilizing patch prediction aggregation for WSI-level classification. Experiments using 5-fold cross-validation demonstrate that our method achieves 96.47 % accuracy and reduces the FID score to 45.72, significantly outperforming class-conditioned baselines.
DiA-gnostic VLVAE: Disentangled Alignment-Constrained Vision Language Variational AutoEncoder for Robust Radiology Reporting with Missing Modalities
Nov 08, 2025Abstract:The integration of medical images with clinical context is essential for generating accurate and clinically interpretable radiology reports. However, current automated methods often rely on resource-heavy Large Language Models (LLMs) or static knowledge graphs and struggle with two fundamental challenges in real-world clinical data: (1) missing modalities, such as incomplete clinical context , and (2) feature entanglement, where mixed modality-specific and shared information leads to suboptimal fusion and clinically unfaithful hallucinated findings. To address these challenges, we propose the DiA-gnostic VLVAE, which achieves robust radiology reporting through Disentangled Alignment. Our framework is designed to be resilient to missing modalities by disentangling shared and modality-specific features using a Mixture-of-Experts (MoE) based Vision-Language Variational Autoencoder (VLVAE). A constrained optimization objective enforces orthogonality and alignment between these latent representations to prevent suboptimal fusion. A compact LLaMA-X decoder then uses these disentangled representations to generate reports efficiently. On the IU X-Ray and MIMIC-CXR datasets, DiA has achieved competetive BLEU@4 scores of 0.266 and 0.134, respectively. Experimental results show that the proposed method significantly outperforms state-of-the-art models.
Unified Cross-Modal Attention-Mixer Based Structural-Functional Connectomics Fusion for Neuropsychiatric Disorder Diagnosis
May 21, 2025Abstract:Gaining insights into the structural and functional mechanisms of the brain has been a longstanding focus in neuroscience research, particularly in the context of understanding and treating neuropsychiatric disorders such as Schizophrenia (SZ). Nevertheless, most of the traditional multimodal deep learning approaches fail to fully leverage the complementary characteristics of structural and functional connectomics data to enhance diagnostic performance. To address this issue, we proposed ConneX, a multimodal fusion method that integrates cross-attention mechanism and multilayer perceptron (MLP)-Mixer for refined feature fusion. Modality-specific backbone graph neural networks (GNNs) were firstly employed to obtain feature representation for each modality. A unified cross-modal attention network was then introduced to fuse these embeddings by capturing intra- and inter-modal interactions, while MLP-Mixer layers refined global and local features, leveraging higher-order dependencies for end-to-end classification with a multi-head joint loss. Extensive evaluations demonstrated improved performance on two distinct clinical datasets, highlighting the robustness of our proposed framework.
Physics-Guided Multi-View Graph Neural Network for Schizophrenia Classification via Structural-Functional Coupling
May 21, 2025Abstract:Clinical studies reveal disruptions in brain structural connectivity (SC) and functional connectivity (FC) in neuropsychiatric disorders such as schizophrenia (SZ). Traditional approaches might rely solely on SC due to limited functional data availability, hindering comprehension of cognitive and behavioral impairments in individuals with SZ by neglecting the intricate SC-FC interrelationship. To tackle the challenge, we propose a novel physics-guided deep learning framework that leverages a neural oscillation model to describe the dynamics of a collection of interconnected neural oscillators, which operate via nerve fibers dispersed across the brain's structure. Our proposed framework utilizes SC to simultaneously generate FC by learning SC-FC coupling from a system dynamics perspective. Additionally, it employs a novel multi-view graph neural network (GNN) with a joint loss to perform correlation-based SC-FC fusion and classification of individuals with SZ. Experiments conducted on a clinical dataset exhibited improved performance, demonstrating the robustness of our proposed approach.
Breast Cancer Classification in Deep Ultraviolet Fluorescence Images Using a Patch-Level Vision Transformer Framework
May 12, 2025Abstract:Breast-conserving surgery (BCS) aims to completely remove malignant lesions while maximizing healthy tissue preservation. Intraoperative margin assessment is essential to achieve a balance between thorough cancer resection and tissue conservation. A deep ultraviolet fluorescence scanning microscope (DUV-FSM) enables rapid acquisition of whole surface images (WSIs) for excised tissue, providing contrast between malignant and normal tissues. However, breast cancer classification with DUV WSIs is challenged by high resolutions and complex histopathological features. This study introduces a DUV WSI classification framework using a patch-level vision transformer (ViT) model, capturing local and global features. Grad-CAM++ saliency weighting highlights relevant spatial regions, enhances result interpretability, and improves diagnostic accuracy for benign and malignant tissue classification. A comprehensive 5-fold cross-validation demonstrates the proposed approach significantly outperforms conventional deep learning methods, achieving a classification accuracy of 98.33%.
Dynamic Contextual Attention Network: Transforming Spatial Representations into Adaptive Insights for Endoscopic Polyp Diagnosis
Apr 28, 2025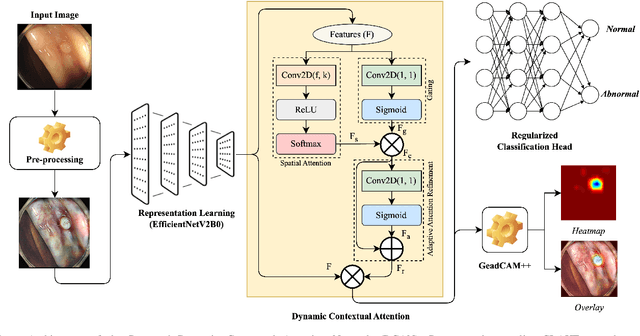
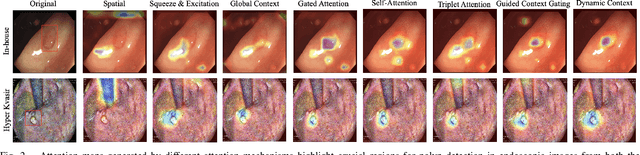
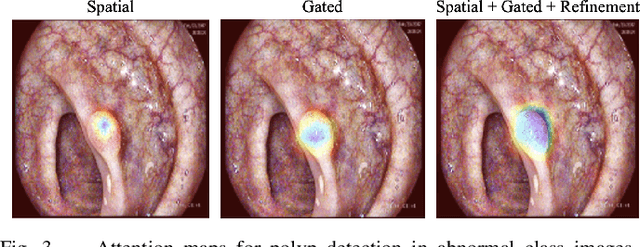
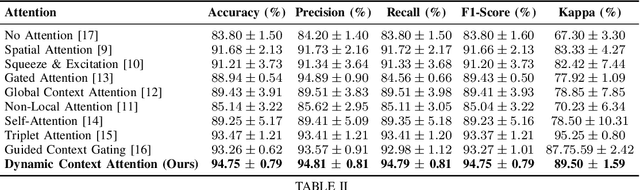
Abstract:Colorectal polyps are key indicators for early detection of colorectal cancer. However, traditional endoscopic imaging often struggles with accurate polyp localization and lacks comprehensive contextual awareness, which can limit the explainability of diagnoses. To address these issues, we propose the Dynamic Contextual Attention Network (DCAN). This novel approach transforms spatial representations into adaptive contextual insights, using an attention mechanism that enhances focus on critical polyp regions without explicit localization modules. By integrating contextual awareness into the classification process, DCAN improves decision interpretability and overall diagnostic performance. This advancement in imaging could lead to more reliable colorectal cancer detection, enabling better patient outcomes.
GCS-M3VLT: Guided Context Self-Attention based Multi-modal Medical Vision Language Transformer for Retinal Image Captioning
Dec 23, 2024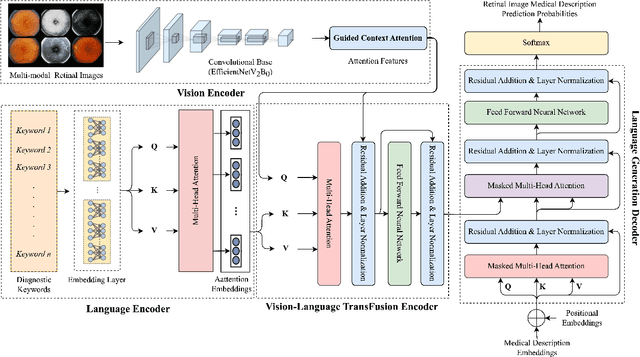
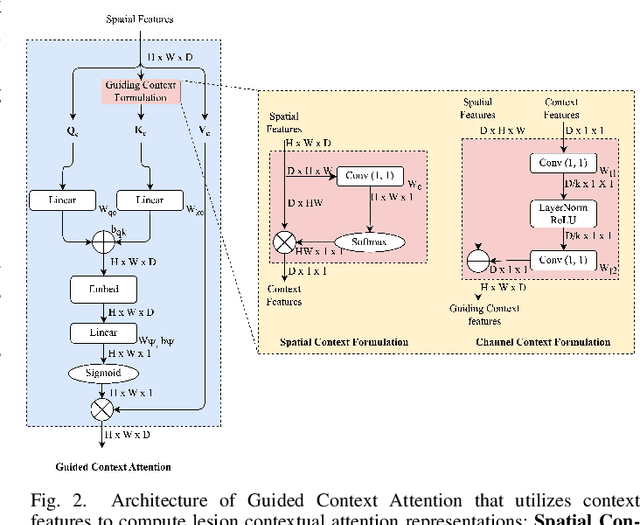
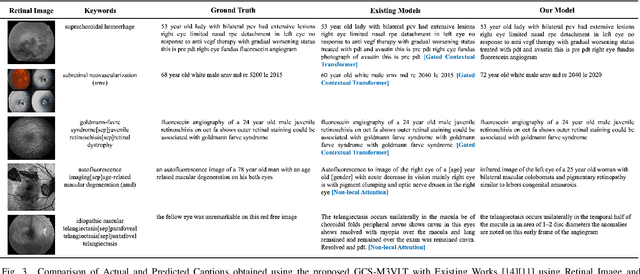
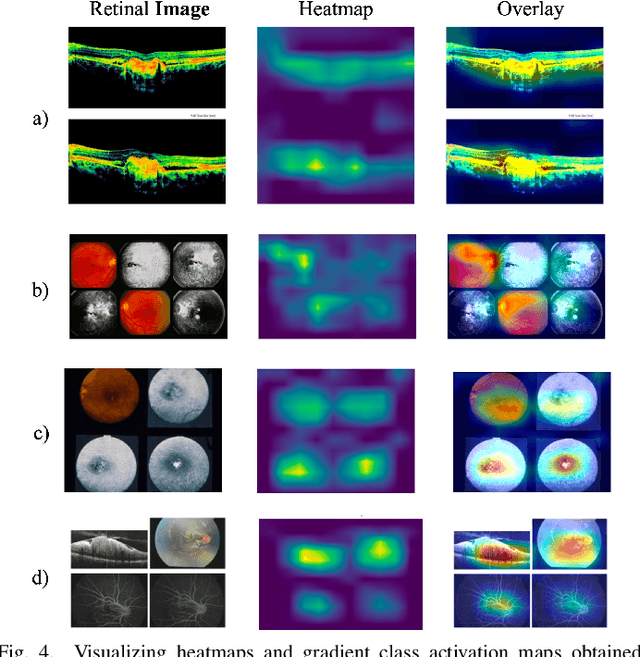
Abstract:Retinal image analysis is crucial for diagnosing and treating eye diseases, yet generating accurate medical reports from images remains challenging due to variability in image quality and pathology, especially with limited labeled data. Previous Transformer-based models struggled to integrate visual and textual information under limited supervision. In response, we propose a novel vision-language model for retinal image captioning that combines visual and textual features through a guided context self-attention mechanism. This approach captures both intricate details and the global clinical context, even in data-scarce scenarios. Extensive experiments on the DeepEyeNet dataset demonstrate a 0.023 BLEU@4 improvement, along with significant qualitative advancements, highlighting the effectiveness of our model in generating comprehensive medical captions.
Multi-modal Imaging Genomics Transformer: Attentive Integration of Imaging with Genomic Biomarkers for Schizophrenia Classification
Jul 28, 2024


Abstract:Schizophrenia (SZ) is a severe brain disorder marked by diverse cognitive impairments, abnormalities in brain structure, function, and genetic factors. Its complex symptoms and overlap with other psychiatric conditions challenge traditional diagnostic methods, necessitating advanced systems to improve precision. Existing research studies have mostly focused on imaging data, such as structural and functional MRI, for SZ diagnosis. There has been less focus on the integration of genomic features despite their potential in identifying heritable SZ traits. In this study, we introduce a Multi-modal Imaging Genomics Transformer (MIGTrans), that attentively integrates genomics with structural and functional imaging data to capture SZ-related neuroanatomical and connectome abnormalities. MIGTrans demonstrated improved SZ classification performance with an accuracy of 86.05% (+/- 0.02), offering clear interpretations and identifying significant genomic locations and brain morphological/connectivity patterns associated with SZ.
Deep learning for automated detection of breast cancer in deep ultraviolet fluorescence images with diffusion probabilistic model
Jul 01, 2024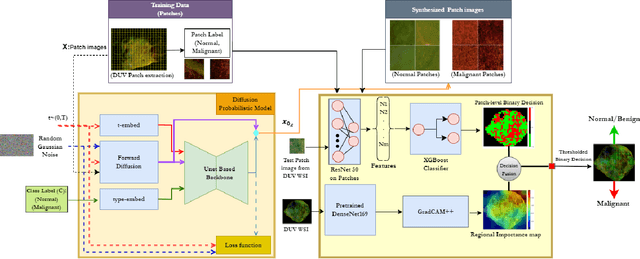
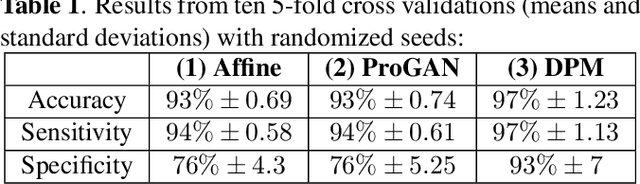

Abstract:Data limitation is a significant challenge in applying deep learning to medical images. Recently, the diffusion probabilistic model (DPM) has shown the potential to generate high-quality images by converting Gaussian random noise into realistic images. In this paper, we apply the DPM to augment the deep ultraviolet fluorescence (DUV) image dataset with an aim to improve breast cancer classification for intraoperative margin assessment. For classification, we divide the whole surface DUV image into small patches and extract convolutional features for each patch by utilizing the pre-trained ResNet. Then, we feed them into an XGBoost classifier for patch-level decisions and then fuse them with a regional importance map computed by Grad-CAM++ for whole surface-level prediction. Our experimental results show that augmenting the training dataset with the DPM significantly improves breast cancer detection performance in DUV images, increasing accuracy from 93% to 97%, compared to using Affine transformations and ProGAN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge