Chuan Qiu
Multi-View Variational Autoencoder for Missing Value Imputation in Untargeted Metabolomics
Oct 12, 2023


Abstract:Background: Missing data is a common challenge in mass spectrometry-based metabolomics, which can lead to biased and incomplete analyses. The integration of whole-genome sequencing (WGS) data with metabolomics data has emerged as a promising approach to enhance the accuracy of data imputation in metabolomics studies. Method: In this study, we propose a novel method that leverages the information from WGS data and reference metabolites to impute unknown metabolites. Our approach utilizes a multi-view variational autoencoder to jointly model the burden score, polygenetic risk score (PGS), and linkage disequilibrium (LD) pruned single nucleotide polymorphisms (SNPs) for feature extraction and missing metabolomics data imputation. By learning the latent representations of both omics data, our method can effectively impute missing metabolomics values based on genomic information. Results: We evaluate the performance of our method on empirical metabolomics datasets with missing values and demonstrate its superiority compared to conventional imputation techniques. Using 35 template metabolites derived burden scores, PGS and LD-pruned SNPs, the proposed methods achieved r2-scores > 0.01 for 71.55% of metabolites. Conclusion: The integration of WGS data in metabolomics imputation not only improves data completeness but also enhances downstream analyses, paving the way for more comprehensive and accurate investigations of metabolic pathways and disease associations. Our findings offer valuable insights into the potential benefits of utilizing WGS data for metabolomics data imputation and underscore the importance of leveraging multi-modal data integration in precision medicine research.
Multi-view information fusion using multi-view variational autoencoders to predict proximal femoral strength
Oct 03, 2022
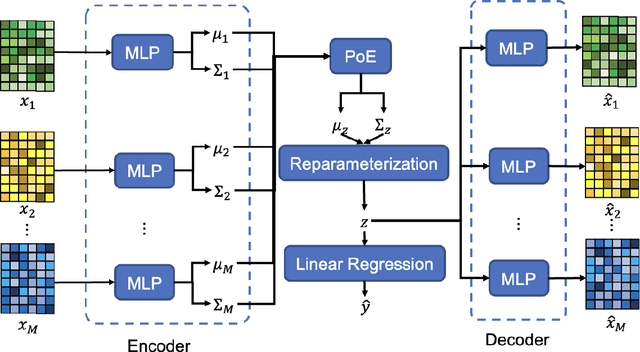
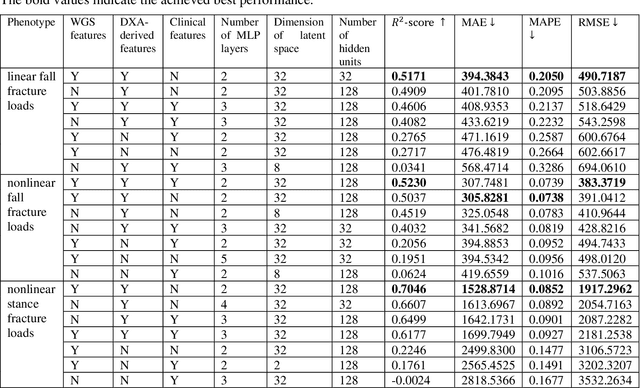
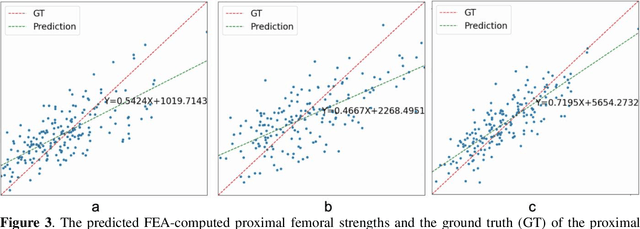
Abstract:Background and aim: Hip fracture can be devastating. The proximal femoral strength can be computed by subject-specific finite element (FE) analysis (FEA) using quantitative CT images. The aim of this paper is to design a deep learning-based model for hip fracture prediction with multi-view information fusion. Method: We developed a multi-view variational autoencoder (MMVAE) for feature representation learning and designed the product of expert model (PoE) for multi-view information fusion.We performed genome-wide association studies (GWAS) to select the most relevant genetic features with proximal femoral strengths and integrated genetic features with DXA-derived imaging features and clinical variables for proximal femoral strength prediction. Results: The designed model achieved the mean absolute percentage error of 0.2050,0.0739 and 0.0852 for linear fall, nonlinear fall and nonlinear stance fracture load prediction, respectively. For linear fall and nonlinear stance fracture load prediction, integrating genetic and DXA-derived imaging features were beneficial; while for nonlinear fall fracture load prediction, integrating genetic features, DXA-derived imaging features as well as clinical variables, the model achieved the best performance. Conclusion: The proposed model is capable of predicting proximal femoral strengths using genetic features, DXA-derived imaging features as well as clinical variables. Compared to performing FEA using QCT images to calculate proximal femoral strengths, the presented method is time-efficient and cost effective, and radiation dosage is limited. From the technique perspective, the final models can be applied to other multi-view information integration tasks.
A generalized kernel machine approach to identify higher-order composite effects in multi-view datasets
Apr 29, 2020



Abstract:In recent years, a comprehensive study of multi-view datasets (e.g., multi-omics and imaging scans) has been a focus and forefront in biomedical research. State-of-the-art biomedical technologies are enabling us to collect multi-view biomedical datasets for the study of complex diseases. While all the views of data tend to explore complementary information of a disease, multi-view data analysis with complex interactions is challenging for a deeper and holistic understanding of biological systems. In this paper, we propose a novel generalized kernel machine approach to identify higher-order composite effects in multi-view biomedical datasets. This generalized semi-parametric (a mixed-effect linear model) approach includes the marginal and joint Hadamard product of features from different views of data. The proposed kernel machine approach considers multi-view data as predictor variables to allow more thorough and comprehensive modeling of a complex trait. The proposed method can be applied to the study of any disease model, where multi-view datasets are available. We applied our approach to both synthesized datasets and real multi-view datasets from adolescence brain development and osteoporosis study, including an imaging scan dataset and five omics datasets. Our experiments demonstrate that the proposed method can effectively identify higher-order composite effects and suggest that corresponding features (genes, region of interests, and chemical taxonomies) function in a concerted effort. We show that the proposed method is more generalizable than existing ones.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge